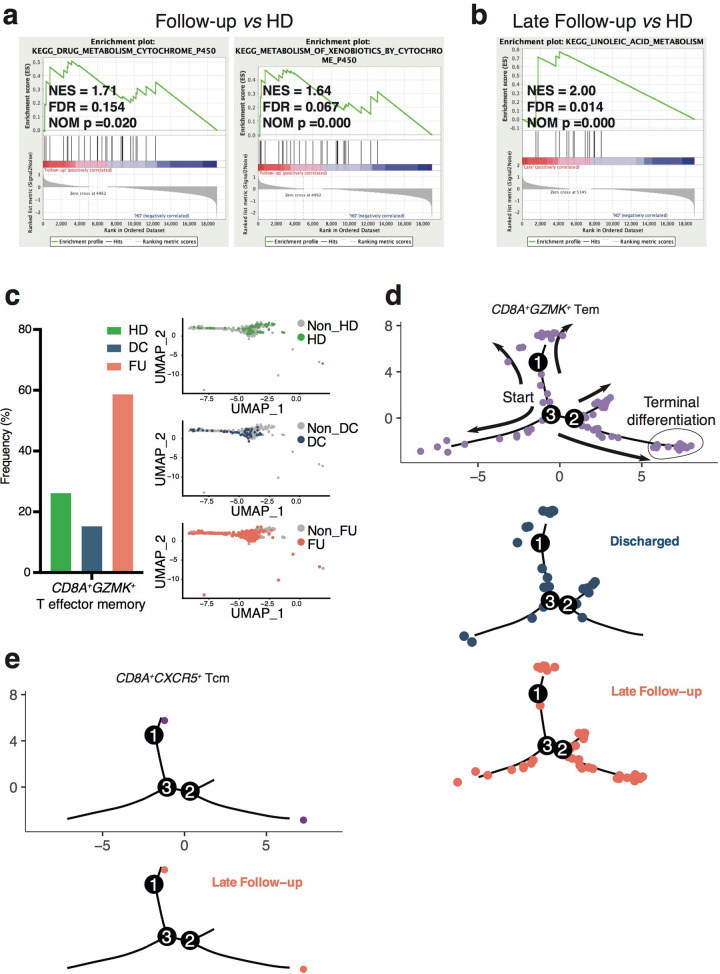
Fig. 6. GSEA for CD3E+CD8A+CD4− clusters of top 20 TCR clonotypes paired scRNA-seq data from Follow-up group vs. HD group and pseudo-time analysis of paired samples from Discharged and Late Follow-up groups.
a Using GSEA to analyze expressed genes from CD3E+CD8A+CD4− clusters of top 20 TCR paired scRNA-seq data, 2 metabolism-related gene sets significantly upregulated in Follow-up group comparing with HD group (n = 7 in Follow-up group, n = 6 in HD group), and b a fatty acid metabolic gene set significantly upregulated in Late Follow-up group vs. HD group analysis (n = 4 in Late Follow-up group, n = 6 in HD group). NES, normalized ES; FDR, false discovery rate; NOM p, normalized p value. c Distribution of each group in CD8A+GZMK+ T effector memory subset. d Pseudo-time analysis of top 20 T cell clones from Discharged and Late Follow-up paired samples. Upper panel, CD8A+GZMK+ Tem subset of combined Discharged and Late Follow-up samples. The arrows indicate the cells’ differentiation trajectories from the beginning to the end; Lower panel, separated Discharged and Late Follow-up groups in CD8A+GZMK+ Tem subset (n = 3 per group). e Pseudo-time analysis of top 20 T cell clones from Discharged and Late Follow-up paired samples. Left panel, distribution of CD8A+CXCR5+ Tcm subset; right panel, only the Late Follow-up samples contributed in CD8A+CXCR5+ Tcm subset (n = 3 per group). DC, Discharged group; FU, Follow-up group; HD, Healthy Donor group; Tcm, central memory T cells; Tem, effector memory T cells.