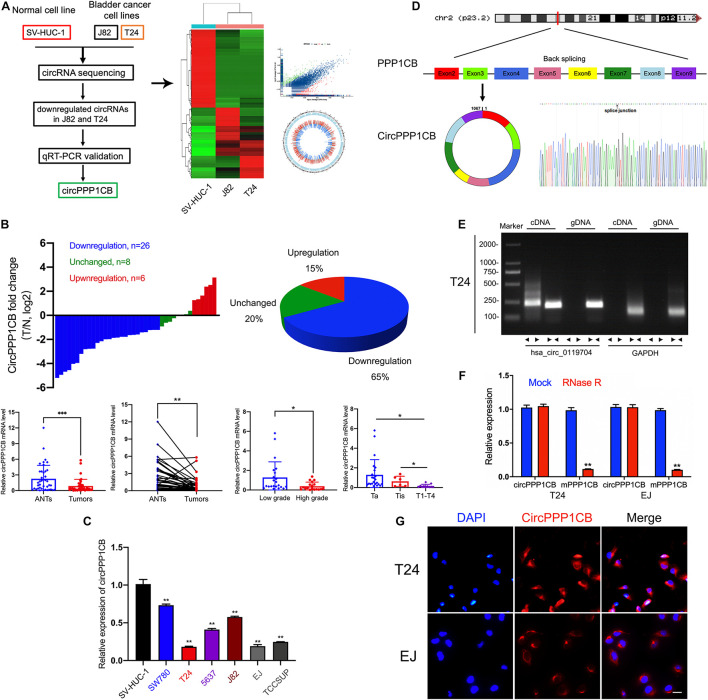
FIGURE 1.
CircRNA sequencing and validation of circPPP1CB in bladder cancer tissues and cells. (A) CircRNA sequencing was performed in SV-HUC-1, J82, and T24 cell lines. A flow diagram, a heat map, and a volcano plot were presented. (B) CircPPP1CB levels were detected in surgical specimens collected from bladder cancer patients (n = 40). CircPPP1CB was downregulated in bladder cancer and was correlated with clinical stages and histological grades. (C). Detection of circPPP1CB fold change in SV-HUC-1, SW780, T24, 5637, J82, EJ, and TCCSUP was performed by qRT-PCR. (D) CircPPP1CB was formed via circularization of exons 2, 3, 4, 5, 6, 7, 8, and 9 of PPP1CB. The head-to-tail splicing junction was also validated and indicated by Sanger sequencing. (E) CircPPP1CB could be amplified efficiently by divergent primers in cDNA in T24 cells. GAPDH was used as a negative control. (F) To verify the tolerance to RNase R, expression levels of circular and linear PPP1BC mRNAs were determined by qRT-PCR. (G) CircPPP1CB was predominantly located in the cytoplasm of bladder cancer cells. CircPPP1CB was stained by a specific probe (red), while nuclei were stained with DAPI (blue). Scale bars, 100 μm. Data are presented as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 vs. control group, ***p < 0.001.