Abstract
Citrus nucellar poly-embryony (NPE) is a mode of sporophytic apomixis that asexual embryos formed in the seed through adventitious embryogenesis from the somatic nucellar cells. NPE allows clonal propagation of rootstocks, but it impedes citrus cross breeding. To understand the cellular processes involved in NPE initiation, we profiled the transcriptomes and DNA methylomes in laser microdissection captured citrus apomictic cells. In apomictic cells, ribosome biogenesis and protein degradation were activated, whereas auxin polar transport was repressed. Reactive oxygen species (ROS) accumulated in the poly-embryonic ovules, and response to oxidative stress was provoked. The global DNA methylation level, especially that of CHH context, was decreased, whereas the methylation level of the NPE-controlling key gene CitRWP was increased. A C2H2 domain-containing transcription factor gene and CitRWP co-expressed specifically in apomictic cells may coordinate to initiate NPE. The activated embryogenic development and callose deposition processes indicated embryogenic fate of nucellar embryo initial (NEI) cells. In our working model for citrus NPE initiation, DNA hyper-methylation may activate transcription of CitRWP, which increases C2H2 expression and ROS accumulation, triggers epigenetic regulation and regulates cell fate transition and NEI cell identity in the apomictic cells.
Keywords: citrus, apomixis, laser microdissection, RNA-Seq, BS-Seq
1. Introduction
In apomictic plants, asexual embryos form without the meiosis and gamete fusion that are required for sexual reproduction. The asexual embryo develops from an unreduced gamete within an unreduced embryo sac that is produced through mitosis (gametophytic apomixis, GA) or directly from somatic cells surrounding the developing sexual embryo sac (sporophytic apomixis, SA). Apomixis has been observed in at least 400 plant species, but rarely reported in crops with the exception of citrus, apple and mango. Citrus use a SA process called nucellar embryogenesis to form poly-embryonic seeds. In this SA process, nucellar cells surrounding the embryo sac spontaneously develop into asexual embryos alongside the zygotic embryo. However, the number of embryos per seed differs among poly-embryonic citrus genotypes, from no more than two [e.g. lemon (Citruslimon)] to as many as 30–40 [e.g. fertilized Satsuma mandarin (C. unshiu)]. The nucellar embryos are genetically identical to the maternal plant and give rise to clonal seedlings for rootstock production and somatic mutants that are potentially useful for scion improvement. On the other hand, nucellar poly-embryony (NPE) is an obstacle to conventional cross breeding because during ovule development, the zygotic embryo may abort because of competition from the nucellar embryos for nutrients from the endosperm. Despite the importance of NPE to citrus propagation and breeding, the mechanism of NPE remains to be elucidated.
GA has been well studied in herbaceous apomictic plants, with the major genetic loci localized and the potential key genes identified.1 Only one gene (i.e. PsASGR-BABY BOOM-like) was found to be a functional regulator of an essential step in GA (i.e. parthenogenesis).2 Asexual propagation can be synthesized either by ectopic expression of BBM and substitution of mitosis for meiosis3 or by editing of the MATRILINEAL (MTL) gene (involved in fertilization) and meiotic genes4 to fix hybrid vigour and stabilize superior heterozygous genotypes of crops. Microarrays and next-generation sequencing have been used to identify differentially expressed genes (DEGs) in the ovules of sexual relative to GA accessions.5,6 In recent years, the apomictic cells, such as apomictic initial (AI) cells and early aposporous embryo sacs, were collected using laser microdissection (LMD) and differences in the transcriptomes that are related to GA were accurately profiled using RNA-Seq.7–9 Epigenetic regulation is also thought to promote GA because GA-like phenotypes were observed in loss-of-function mutants with deficiencies in DNA methyltransferases and the small RNA mediated RNA-directed DNA methylation (RdDM) pathway in maize and Arabidopsis.10–12
Genetic localization of genes associated with NPE in citrus has progressed in recent years. The NPE locus in Citrus genus was initially mapped within a 380-kb region on chromosome 4.13 Molecular markers tightly linked to NPE were also developed in the Poncirus genus.14 The candidate NPE-controlling gene CitRWP was identified from a more accurately defined 80-kb locus that was based on BSA and GWAS analysis.15 Antisense suppression of CitRKD1 (i.e. CitRWP) abolished NPE in sweet orange and thus demonstrated a key role for CitRWP in citrus NPE.16 However, the regulatory networks embedded in the cell fate transition and the specification that occurs during the initiation of NPE remains to be uncovered.
Expressed sequences were previously compared in the ovules of mono- and poly-embryonic citrus varieties.17,18 We previously profiled DEGs and miRNAs in ovules from two pairs of mono- and poly-embryonic citrus varieties.19 Questions remain, however, about the processes that occur specifically in the apomictic cells embedded in nucellar tissue. Here, we describe genome-wide gene activity and DNA methylation landscapes in citrus apomictic cells during different stages of nucellar embryo initial (NEI) cell development using LMD, whole-genome RNA-Seq and bisulfite sequencing (BS-Seq). We gained new insights into the initiation of NPE in citrus and SA in plants.
2. Materials and methods
2.1. Plant materials
For histological observation of nucellar embryo development, we collected ovaries from mono-embryonic Clementine mandarin and poly-embryonic Valencia sweet orange for the preparation of paraffin sections at different stages [3 days before anthesis (DBA); 0 day after anthesis (DAA); 3 DAA; 7 DAA; and every 7 days until seed maturation]. Ovaries of another six poly-embryonic varieties were collected and sectioned at 7 DAA. The six varieties were the F1 hybrid (code 194) of ‘Hirado Buntan’ pummelo (mono-embryonic) × Fairchild mandarin (poly-embryonic) (HB×FC cross), ‘Guoqing No.1’ Satsuma mandarin, Red tangerine, ‘Cocktail’ grapefruit, ‘Willow leaf’ mandarin and ‘Huagan No.2’ Ponkan mandarin. The NPE capacity was quantified by calculating the average number of embryos per seed. The variety with NEI cells that were distinguishable by observing unstained paraffin sections with a microscope was chosen for LMD capturing.
At an earlier stage of NEI cell development, when the sexual embryo sac is undergoing mega-gametogenesis and the NEI cells are not observable under a microscope, ovaries were harvested separately at 3 DBA from 10 mono-embryonic and 10 poly-embryonic progeny from the segregating F1 population derived from the ‘HB×FC’ cross (Supplementary Table S1). One biological replicate contained ovaries that were harvested from five trees and then combined. Two biological replicates were used for each genotype. At a later stage of NEI cell development, when the zygote in the embryo sac is undergoing mitosis and the NEI cells are observable under a microscope, ovules were harvested 7 DAA from four trees of the highly poly-embryonic Ponkan mandarin (C. reticulata Blanco) variety ‘Huagan No.2’ (Supplementary Table S1). One biological replicate contained ovaries that were harvested from two trees and combined. Two biological replicates were also used for each cell type at a later stage of NEI cell development. The adult trees were all grown at the Institute of Citrus Science, Huazhong Agricultural University (Wuhan, China).
2.2. Capturing of apomictic cells and extraction of DNA/RNA
Paraffin section preparation, LMD capturing and the extraction of DNA and RNA from the captured cells were conducted as previously described.20 The specific cell lineages were captured from the 8–10 μm paraffin sections of ovaries using a LMD7000 system (Leica Microsystems). To ensure the consistency of each replicate, equal areas of cells were captured from the ovary sections from each tree for each replicate. RNA was exacted from the LMD captured sections using the Arcturus PicoPure RNA Isolation Kit (Life Technologies) and treated with the RNase-Free DNase Set (Qiagen). DNA was exacted using the QIAamp DNA FFPE Tissue Kit (Qiagen) and treated with RNase A (Invitrogen).
2.3. Construction of RNA-Seq and BS-Seq libraries
RNA quality was examined using the Agilent RNA 6000 Pico chip and the 2100 Bioanalyzer (Agilent) (Supplementary Table S2). DNA quality was examined using the Agilent High Sensitivity DNA Kit (Supplementary Table S3). RNA was exacted from an area of 10.7–17.8 × 106 μm2 LMD-captured sections for each replicate and converted to first-strand cDNA using the SMARTer® Ultra™ Low RNA Kit for Illumina® Sequencing (Clontech) before RNA-Seq library preparation. Approximately 100 ng of DNA combined with lambda DNA (the negative control) was fragmented and ligated to the cytosine-methylated adapters before bisulfite conversion using the EZ DNA Methylation Gold Kit (Zymo Research). The libraries were sequenced on the Illumina Hiseq 2000 platform (Novogene, Tianjin, China).
2.4. Analysis of the RNA-Seq and BS-Seq data
Over 91.6% of the high-quality reads were mapped to the citrus genomes,15,21 with no mismatch allowed, and assembled into transcripts using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts) and StringTie22 (Supplementary Table S4). Correlation coefficients were calculated based on transcript abundance that was normalized to fragments per kb of exon per million fragments mapped (FPKM) (Supplementary Table S5). The DEGs were profiled using edgeR23 with a false discovery rate (FDR) < 0.05 and a fold-change ≥ 2. Gene ontologies (GO) were assigned and enriched (FDR < 0.05) using the web-based agriGO programme.24
The high-quality reads of BS-Seq were mapped to the citrus genome with Bismark (v0.18.0; -N 1, -L 20).25 The bisulfite non-conversion rate (0.021–0.025) was estimated based on the bisulfite conversion rate of the control lambda DNA (Supplementary Table S6). The sequencing-error frequency (0.001–0.008) was estimated by the proportion of methylated Cs in the Cs uniquely mapped to the mitochondrial genome26 (Supplementary Table S7). The methylated cytosines (mCs) and differentially methylated regions (DMRs) were identified as previously described.27 Differentially methylated genes (DMGs) were defined as gene bodies or 2 kb promoter regions that overlapped with DMRs.
2.5. DNA methylation assay and 5-azacytidine treatment of citrus
Genomic DNAs were isolated from the ovules of poly-embryonic (Ponkan) and mono-embryonic (Clementine mandarin) at an earlier stage of NEI cell development and bisulfite treated using Methylation Gold kit (Zymo Research). Treated DNAs were analysed using PCR with ZymoTaq PreMix. The PCR products were ligated into the pTOPO-Blunt vector. Sequences of 30 independent clones for each fragment were analysed using Kismeth.28 The primers used for BS-Seq are listed in Supplementary Table S8.
We sprayed 2-year-old poly-embryonic mini-citrus (Fortunella hindsii) grown in greenhouse with a 4 mM solution of the DNA methylation inhibitor, 5-azacytidine (5′-Aza). Three plants were sprayed twice a day for 10 days. Young leaves were collected for RNA extraction and quantification of relative gene expression levels using real-time quantitative PCR (qRT-PCR). Another three plants were sprayed with water and used as the controls.
2.6. qRT-PCR analysis
First-strand cDNA was synthesized using RNA isolated using the LMD procedure and the oligo-dT30VN primer followed by second-strand cDNA synthesis. Total RNA was extracted from ovules using the TRIzol reagent (Takara, Dalian) and reverse-transcribed using HiScript II QRT SuperMix for qPCR (Vazyme). Real-time PCR was conducted and analysed as previously described.29 Gene-specific qRT-PCR primers are listed in Supplementary Table S8. CsActin was used as the internal control.
2.7. RNA in situ hybridization
RNA in situ hybridization experiments were conducted as described previously.30 The 10-μm thick paraffin sections were mounted onto slides. Each slide contained an equal number of poly- and mono-embryonic ovary sections at the same developmental stage. Primers used for the PCR-based synthesis of RNA probes are listed in Supplementary Table S8.
2.8. Subcellular localization of protein
The C2H2 (Cg1g000740) CDS without a stop codon was amplified using sequence-specific primers (Supplementary Table S8) and inserted into p2GWY7-35S to generate the 35S::C2H2-yellow fluorescent protein (YFP) fusion construct. The nuclear localized rice OsGhd7 was used as a nuclear marker and was expressed from the 35S::OsGhd7-cyan fluorescent protein (CFP) transgene.31 Citrus mesophyll protoplasts were prepared and co-transformed with constructs that express the target and the nuclear marker fused to fluorescent proteins. The images were acquired using a confocal laser-scanning microscope (Leica TCS SP2, Germany).
2.9. Detection of reactive oxygen species and assay of enzyme activity
Superoxide anion (O2–) and hydrogen peroxide (H2O2) in ovules were histochemically stained with nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB), respectively.32 Superoxide dismutase (SODs) and peroxidases (PODs) activities in ovaries was assayed spectrophotometrically after the removal of the ovary walls as described by previously.33
3. Results
3.1. Morphological and cytological characterization of NPE in citrus
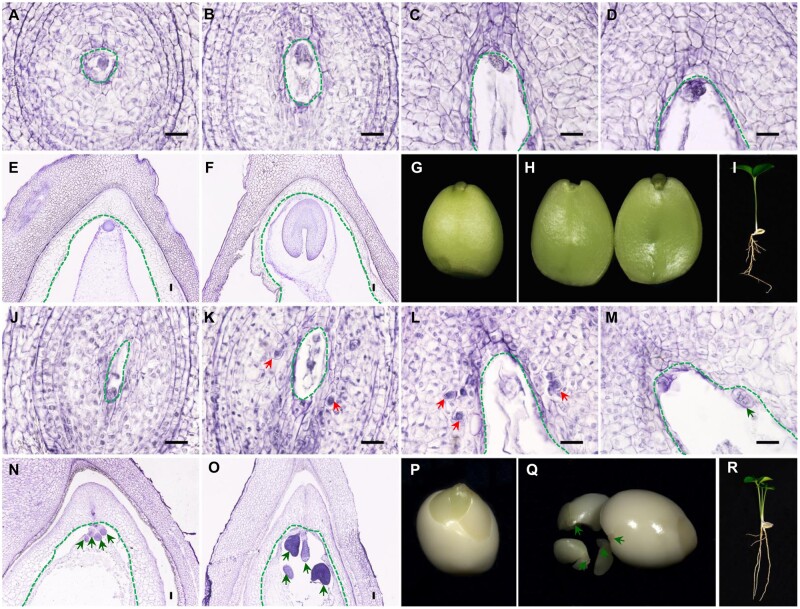
In the ovules of mono-embryonic citrus (Clementine mandarin), the zygotic embryo and endosperm arise from double-fertilization in the mature embryo sac and give rise to one seedling (Fig. 1A–I). By contrast, in the ovules of poly-embryonic citrus (Valencia sweet orange), multiple asexual embryos initiate directly from the nucellar cells and develop alongside the zygotic embryo that is localized in the sexual embryo sac (Fig. 1J–O). In the poly-embryonic ovule, nucellar cells are somehow reprogrammed and turned into NEI cells containing an enlarged nucleus, deeply stained cytoplasm and a thickened cell wall, which are histologically evident in ovules at 7 DAA (Fig. 1K). The sporophytic nucellar cells degrade gradually as ovules develop. The NEI cells, however, are protected from degradation (Fig. 1L) and instead undergo mitosis that directs them toward embryonic development and enter the enlarging embryo sac (Fig. 1M). Multiple nucellar embryos undergo a morphogenesis process that is similar to the zygotic embryo (Fig. 1N and O) and develop into mature asexual embryos (Fig. 1P and Q). The zygotic embryo, however, may abort during development, because of nutritional competition with the asexual embryos in the same embryo sac. Therefore, the germination of one mature ‘Valencia’ seed yields multiple seedlings with no more than one sexual progeny (Fig. 1P–R). We sectioned and observed ovaries from six poly-embryonic citrus varieties with different capacities for NPE (2.5–9.2 embryos per seed) (Supplementary Fig. S1). Ponkan mandarin is highly poly-embryonic (with 9.2 ± 4.4 embryos per seed), thus ovaries from Ponkan mandarin produced plenty of NEI cells that were clustered together and were evident in the unstained ovule sections (Supplementary Fig. S1G). Therefore, Ponkan mandarin was used for capturing the NEI cells.
Figure 1.
Embryo initiation, development and seed germination in mono-embryonic and poly-embryonic citrus. (A–F) Paraffin sections of ovules and seeds from mono-embryonic Clementine mandarin illustrating sexual embryo development. (G–I) Mature mono-embryonic seed of Clementine mandarin with the seed coat removed and the single plantlet germinated from one seed. (J–O) Paraffin sections of ovules and seeds from poly-embryonic Valencia sweet orange. The emergence of the nucellar embryo initial (NEI) cells and the development of nucellar embryos are apparent. (P–R) Mature poly-embryonic seed of Valencia sweet orange with the seed coat removed and multiple plantlets germinated from one seed. The red arrows point to NEI cells. The green arrows point to nucellar poly-embryos. The green dotted lines enclose the sexual embryo sac. Scale bars 20 μm.
3.2. The apomictic and sporophytic cells differ significantly in biological processes
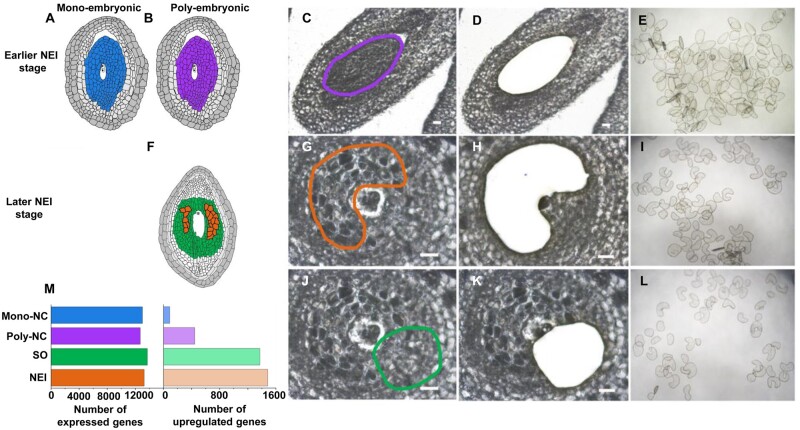
A total of 13,967 and 15,779 genes were detected (FPKM ≥ 1) using RNA-Seq at earlier and later stages of NEI cell development, respectively. The four cell types share similar gene expression level distributions, with similar percentages of genes spanning six orders of magnitude (Supplementary Fig. S2A). We quantified the relative expression of 22 selected genes (Supplementary Fig. S3). The expression patterns that were demonstrated using qRT-PCR were highly consistent with the expression patterns demonstrated using RNA-Seq (r > 0.87).
At an earlier stage of NEI cell development (3 DBA), the reprogramming of the Poly-NC is underway but the NEI cells are not yet formed or observable. In contrast, at this stage of development, the reprogramming of the Mono-NC has not begun. To profile biological processes and genes active during cellular reprogramming, transcriptomes of the 6–7 layers of nucellar tissues captured from the ovary sections (i.e. Poly-NC and Mono-NC) of the F1 progeny from mono- and poly-embryonic citrus were compared (Fig. 2A–E). A total of 532 DEGs were identified. The expression of 444 (83.5%) of these genes was up-regulated in the Poly-NC. The expression of the remaining 88 genes was down-regulated in the Poly-NC (Fig. 2M, Supplementary Table S9). Biological processes that were enriched in the DEGs of the poly-NC and the Mono-NC are listed in Fig. 3A. Notably, the cellular process ‘mitochondrial respiratory chain complex I’ was enriched among the genes that were up-regulated in the Poly-NC (Supplementary Table S10).
Figure 2.
Laser microdissection of apomictic and sporophytic cells and differentially expressed genes (DEGs) profiled by RNA-Seq. (A–E) Schematic diagrams of cell lineages in mono- (A) and poly-embryonic (B) ovules at an earlier stage of NEI cell development. (C–E) Nucellar tissue collected separately from the mono- and poly-embryonic ovules at 3 days before anthesis from the F1 progeny derived from a ‘HB pummelo (mono-embryonic) × Fairchild mandarin (poly-embryonic)’ cross. (f) Schematic of cell lineages in a poly-embryonic ovule at a later stage of NEI cell development (7 days after anthesis). Nucellar embryo initial (NEI) cells (G–I) and nucellar sporophytic (SO) cells (J–L) were captured from Ponkan mandarin ovules. The paraffin sections show ovules before microdissection (C, G and J), after microdissection (D, H and K) and the collected cells (E, I and L). (M) Number of expressed genes and DEGs in specific cell lineages. The earlier and later stages of NEI cell development refer to the stages before and after the appearance of nucellar embryo initial (NEI) cells in the nucellus. Poly/Mono-NC, poly/mono-embryonic nucellar tissue. Scale bars 20 μm.
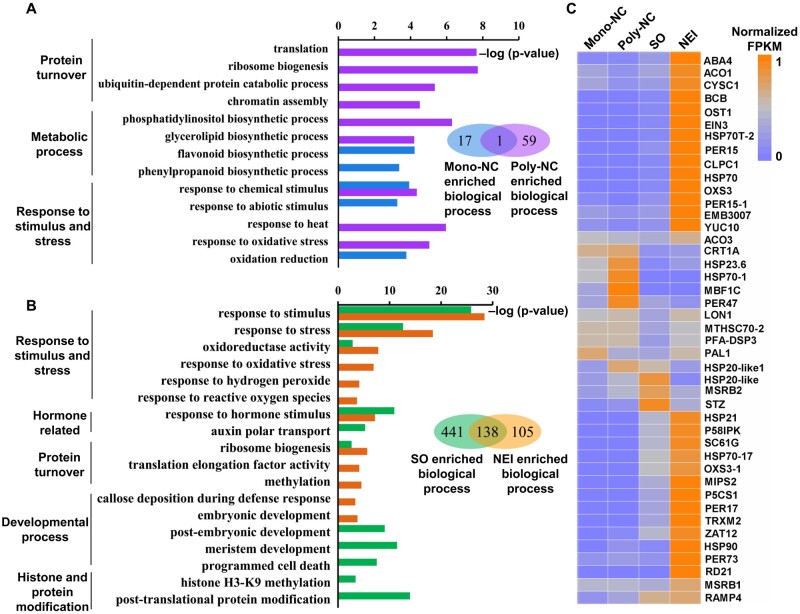
Figure 3.
Analysis of transcriptomes in apomictic and sporophytic cells. Enriched GO terms (biological processes) among differentially expressed genes (DEGs) between apomictic and sporophytic cells at earlier (A) and later (B) stages of NEI cell development. (C) Heatmap of 43 selected genes that are up-regulated in apomictic cells and associated with the GO term ‘response to oxidative stress’. The orange-to-blue scale indicates gene expression levels. orange = most, grey = moderate, blue = least. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
To profile the active genes and biological processes in the NEI cells, transcriptomes were compared between the LMD-captured NEI cells and the surrounding nucellar sporophytic (SO) cells of Ponkan mandarin at later stage of NEI cell development, after the NEI cells enlarged and developed a dense cytoplasm and were observable in ovary sections (7 DAA) (Fig. 2F–L). A total of 2,859 DEGs were identified from a comparison of the transcriptomes of NEI cells and SO cells. We found that the expression of 1,483 (51.9%) genes was up-regulated and that the expression of 1,376 (48.1%) genes was down-regulated in NEI cells (Fig. 2M, Supplementary Table S12). These DEGs are associated with particular biological processes (Fig. 3B).
The biological processes responsible for the production (ribosome biogenesis and translation) and degradation (ubiquitin-dependent protein catabolic) of proteins were associated with the most significantly enriched GO terms for genes that were up-regulated in the Poly-NC (Fig. 3A). NEI cells up-regulated genes were enriched in ribosome biogenesis and methylation process (Fig. 3B). The majority of genes associated with ribosome biogenesis (25/27), translation processes (65/73) and methylation process (30/57) encode different ribosomal proteins. Consistent with this finding, the expression levels of genes associated with ‘ribosome’ and ‘ubiquitin’ were up-regulated in both the Poly-NC and NEI cells (Supplementary Fig. S2B). These results demonstrate that protein turnover (both production and degradation) was activated in the apomictic cells during NEI cell development.
Chromatin assembly process was enriched among genes that were up-regulated in the Poly-NC (Fig. 3A). In contrast, genes associated with the methylation of histone H3-K9 were enriched among genes that were down-regulated in NEI cells (Fig. 3B), including a gene that encodes SUVH4, which catalyses H3 lysine 9 dimethylation. This modification is required for the maintenance of CHG methylation in Arabidopsis.34 The expression of SUVH4 was down-regulated 2.5-fold in NEI cells (FPKM = 8.7). These data provide evidence that the reprogramming of chromatin occurs during NEI cell development. The genes associated with responses to oxidative stress were overrepresented among the DEGs enriched in both the Poly-NC (24 up-regulated genes; Supplementary Table S10) and NEI cells (60 up-regulated genes; Supplementary Table S13) (Fig. 3C). The majority of these genes encode peroxidases (PERs), heat shock proteins and stress-responsive proteins, such as RD21 (Fig. 3C). In contrast, eight genes responsible for the biosynthesis of antioxidant flavonoids and anthocyanins that are associated with phenylpropanoid and flavonoid biosynthetic processes (Supplementary Table S11) were enriched among the genes that were down-regulated in the Poly-NC (Fig. 3A).
Genes associated with embryonic development and callose deposition processes were enriched among the genes that were up-regulated in NEI cells. In contrast, genes associated with these GO terms were enriched among the down-regulated genes during post-embryonic development and meristem development (Fig. 3B, Supplementary Fig. S2C, Supplementary Table S14), which reflects the early embryogenic cell fate of NEI cells.
Phytohormone-related biological processes were not differentially enriched among the DEGs associated with the Poly-NC. In contrast, response to hormone stimulus was enriched among the DEGs associated with the NEI cells and auxin polar transport was enriched among the genes that were down-regulated in NEI cells. We analysed the expression levels of genes associated with the biosynthesis, signalling, and transport of auxin (82 genes), gibberellin (21 genes) and cytokinin (54 genes). Indeed, none of these genes were differentially expressed in the Poly-NC relative to the Mono-NC. However, we were able to detect the expression of 58 genes associated with auxin. Eight of these genes were up-regulated and 16 of these genes were down-regulated in the NEI cells. The down-regulated genes include two genes associated with auxin biosynthesis, 12 genes associated with auxin signalling and 2 genes associated with auxin transport (Supplementary Fig. S4). Among the 21 genes associated with gibberellins, the expression of 6 genes associated with gibberellin biosynthesis was up-regulated in NEI cells (Supplementary Fig. S4).
3.3. Global DNA methylation, especially in the CHH context, was reduced in apomictic cells
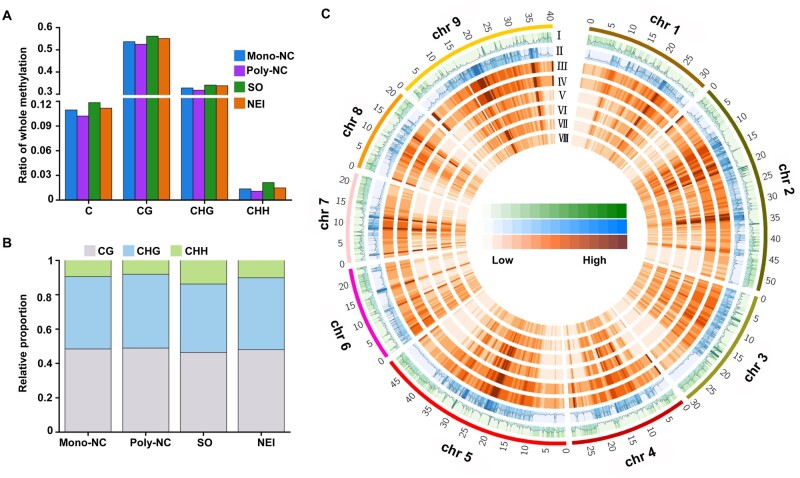
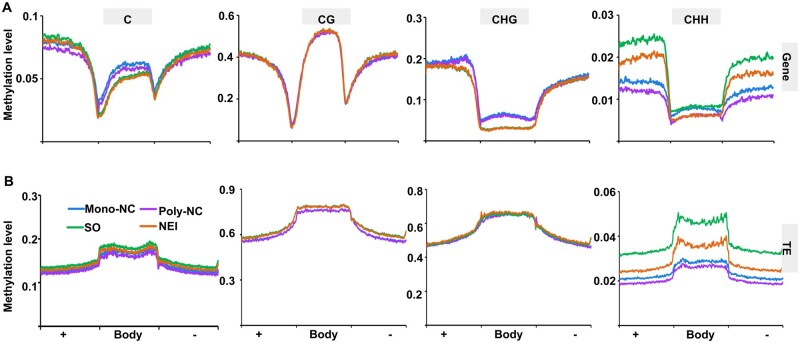
To investigate DNA methylation profiles during NEI cell development, whole-genome BS-Seq was conducted using the same sets of LMD-captured apomictic and sporophytic cells. For the CHH context, we determined that the average CHH methylation level was 1.0–2.1%, based on the identification of ∼0.6–1.3 million (∼8–14% of the total) mCs that contribute to the CHH context (Fig. 4A and B). The DNA methylation levels in the three contexts were all higher in the chromosomal regions that are dense with transposable elements (TEs), relative to regions with high gene density (Fig. 4C; Supplementary Fig. S5A) and were also higher in TEs than in protein-coding genes (PCGs) (Fig. 5). The total levels of C methylation in both PCGs and TEs, especially the levels of CHH methylation, were lower in the apomictic cells than in the sporophytic cells at both stages of NEI cell development (Poly-NC < Mono-NC, NEI cells < SO cells) (Fig. 5).
Figure 4.
Genomic landscape of DNA methylation in apomictic and sporophytic cells at two stages of NEI cell development. (A) Percentages of methylated cytosines (mCs). (B) Ratios of mCs in CG, CHG and CHH contexts. (C) Circos plot showing gene density, TE density, ratios of CG, CHG and CHH methylation in Poly-NC and Mono-NC in 512-kb window among genome. The outer black track represents the nine chromosomes of the pummelo (C. grandis) genome. Tracks I, gene density; Tracks II, TE density; Tracks III–, ratio of methylation in specific C contexts in specific cell lineages, including CG-Mono-NC, CG-Poly-NC, CHG-Mono-NC, CHG-Poly-NC, CHH-Mono-NC, CHH-Poly-NC, respectively. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
Figure 5.
Comparison of DNA methylation levels in the body and flanking regions of genes (A) and TEs (B) during the initiation of nucellar embryogenesis. In all cases, the flanking regions are the same length as the gene body or TE body. +, Body, −, the upstream, body and downstream regions of genes/TEs; methylation levels were calculated in 100 proportionally sized bins. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
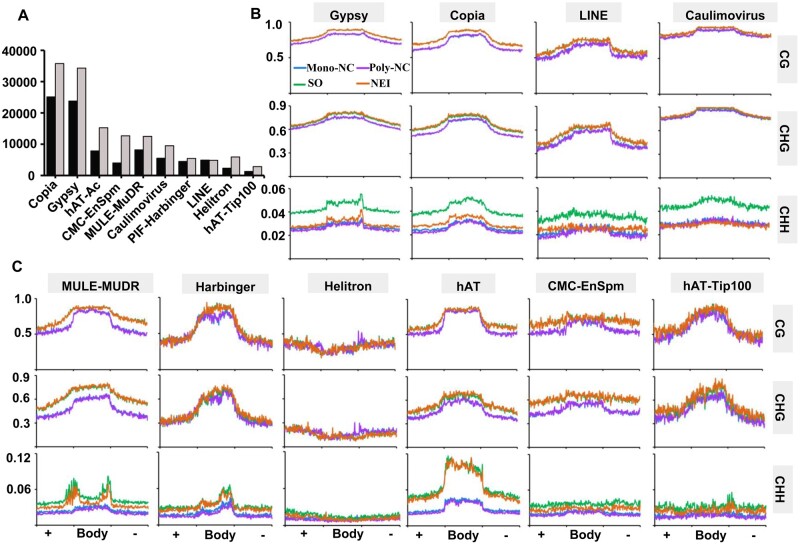
Among the 10 biggest citrus TE groups (Fig. 6A), the CHH methylation levels of the Class I TEs (retrotransposons including Gypsy, Copia, LINE and Caulimovirus) was obviously lower in NEI cells than in SO cells (Fig. 6B). The CHH methylation levels of the Class II TEs (DNA transposons) was also slightly lower in NEI cells (Fig. 6C). The CHH methylation levels of both classes of TEs were slightly lower in the Poly-NC relative to the Mono-NC. The results demonstrated that global DNA methylation levels, especially in the CHH context, were decreased in the apomictic cells and that the reduction in the methylation of NEI cells was primarily due to the reduction of methylation levels of retrotransposons.
Figure 6.
Comparison of DNA methylation levels in different classes of TEs during the initiation of nucellar embryogenesis. (A) Categories and numbers of TEs identified in Clementine mandarin (C. clementina, black columns) and pummelo (C. grandis, grey columns) genomes. Average distribution of DNA methylation levels in Class I (B) and Class II (C) TEs. In all cases, the TE flanking regions are of the same length as the TE bodies. +, Body, −, the upstream, body and downstream regions of TEs; methylation levels were calculated in 100 proportionally sized bins. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
We examined the expression levels of 81 DNA methylation pathway gene homologs, including those involved in RdDM. Among the 62 genes detected at earlier stage of NEI cell development, none were differentially expressed in the Poly-NC relative to the Mono-NC. Among the 57 genes detected at later stage of NEI cell development, the expression of three RdDM pathway genes was up-regulated in NEI cells. In contrast, seven genes were down-regulated in NEI cells, including two genes that encode DNA methyltransferases, one gene that encodes a DNA glycosylase involved in active demethylation [REPRESSOR OF SILENCING 1 (ROS1)] and four RdDM pathway genes (Supplementary Fig. S4).
3.4. DNA methylation may alter expression of apomixis-related genes
To investigate the potential effect of DNA methylation on the initiation of NPE, DMRs, DMGs and differentially methylated TEs were identified in the apomictic cells (Supplementary Fig. S5B, C; Supplementary Tables S15 and S16). The DMGs were subjected to a Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis (Supplementary Fig. S6). Among the Poly-NC hyper-methylated genes, the ribosome pathway was significantly enriched. These findings are consistent with the up-regulated expression of genes associated with ribosome-related processes in the Poly-NC (Fig. 3A; Supplementary Fig. S2B). We suggest that the activated expression of the ribosomal proteins may be associated with the hyper-methylation of DNA. Notably, plant hormones signal transduction pathways were enriched among hypo-methylated genes in NEI cells. Approximately half (19/44) of these genes encode auxin-responsive proteins (Supplementary Fig. S6D). Moreover, the expression of auxin-related pathway genes was down-regulated in NEI cells (Fig. 3B, Supplementary Fig. S4). These data are consistent with an association between the down-regulated expression of auxin-related genes and the hypo-methylation of DNA.
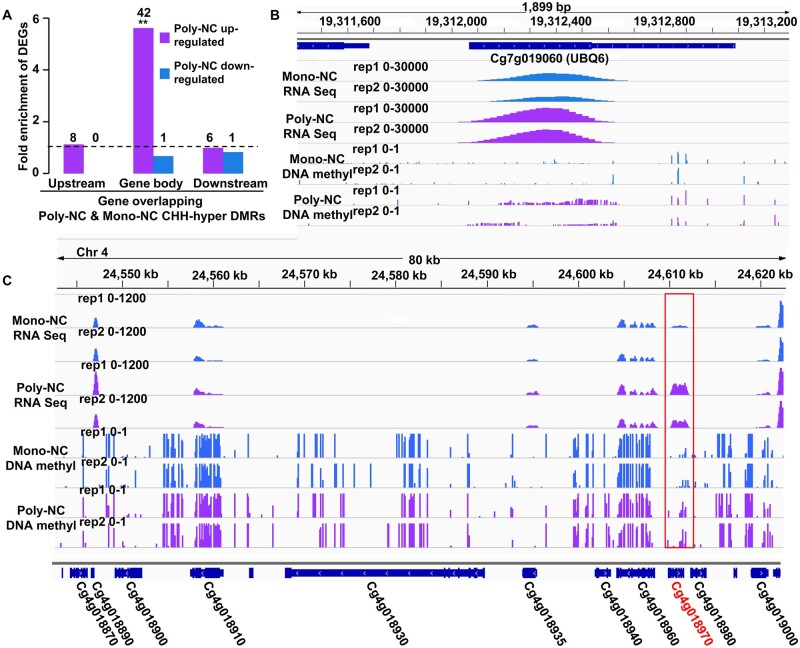
In order to identify DNA methylation regulated genes, DEGs with overlapped DMRs were listed (Supplementary Tables S17 and S18). We compared the DEGs with the DMRs and found that genes that were up-regulated in the Poly-NC were significantly enriched in CHH hyper-methylation in the gene body (Fig. 7A). These data indicate a positive association between CHH methylation of the gene body and gene expression in the Poly-NC. For example, UBIQUITIN EXTENSION PROTEIN 6 (UBQ6), a key regulator of plant development and callogenesis, was up-regulated and had higher levels of DNA methylation in the gene body in the Poly-NC (Fig. 7B). Among the 11 genes within the ∼80-kb citrus NPE locus,15 only CitRWP (Cg4g018970) was up-regulated in apomictic cells (Poly-NC and NEI cells). Additionally, a hyper-methylated DMR was detected in the gene body of CitRWP in the Poly-NC (Fig. 7C). It is known that an NPE-specific miniature inverted-repeat transposable element (MITE) insertion in the promoter region is associated with the transcriptional upregulation of CitRWP for poly-embryonic development.15,16,35 We tested whether the MITE insertion was associated with the hyper-methylation of CitRWP. According to whole genome BS-Seq, a CHH hyper-methylation signal was detected in the downstream region proximal to the MITE in the Poly-NC, but not detected in the same region in the Mono-NC, which lacks the MITE (Supplementary Fig. S7A). In the poly-embryonic Ponkan (with the MITE), the CHH hyper-methylated region was detected in both NEI and SO cells (Supplementary Fig. S8B). Using local BSP-PCR, we determined that the DNA methylation levels of all sequence contexts associated with the MITE (located 5′ to the start codon from –133 to –560 bp) and both the upstream (–716 to –560) and downstream (–133 to 169 bp) proximal regions were significantly higher in the poly-embryonic Ponkan ovules than in the same region (–289 to 169 bp, without the MITE) in the mono-embryonic Clementine mandarin ovules at an earlier stage of NEI cell development (Supplementary Fig. S7B and C). Therefore, we suggest that the insertion of the MITE is associated with the hyper-methylation of the MITE and the proximal region. To test whether DNA methylation affects the expression of CitRWP, we treated the mini-citrus plants with the DNA methylation inhibitor (5′-Aza) and found that the expression of CitRWP was significantly decreased in the treated leaves (Supplementary Fig. S9). Therefore, we suggest that DNA methylation positively regulates the expression of CitRWP.
Figure 7.
Influence of differentially methylated regions (DMRs) on gene expression. (A) Fold-enrichment of DEGs in CHH-hyper DMRs relative to all genes in the Poly-NC relative to the Mono-NC. Fisher’s test was used to infer statistical significance (*P < 0.05; **P < 1e-10). The numbers above the columns indicate DEGs in Poly-NC relative to Mono-NC that overlapped with CHH-hyper DMRs. (B) The genome browser snapshot shows the expression levels and DNA methylation levels of UBQ6 (Cg7g019060). (C) Genome browser snapshot showing the expression levels and CG methylation levels of the 80-kb poly-embryony locus in the Poly-NC and the Mono-NC. The red box indicates a differentially methylated region. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
3.5. Apomictic cell-specific genes in citrus
At an earlier stage of NEI cell development, only 21 transcription factor (TF) mRNAs were differentially expressed in the Poly-NC relative to the Mono-NC. Only 12 of the DEGs were up-regulated in the Poly-NC (Supplementary Fig. S10A and B), including CitRWP. Indeed, the expression of CitRWP was up-regulated 7-fold in the Poly-NC (FPKM = 17). Notably, the expression of an mRNA (Cg1g000740) that encodes a TF with a C2H2 domain (hereafter referred to as C2H2) was up-regulated 45-fold in the Poly-NC (FPKM = 409)—the highest level of induced expression among all DEGs. Among the 212 TF-encoding mRNAs that were differentially expressed in NEI cells relative to SO cells at a later stage of NEI cell development, there are genes that contribute to development and stress responses (Supplementary Fig. S10C and D). Significantly, the expression of CitRWP and C2H2 was also up-regulated in NEI cells. The expression of well-known embryogenesis-related TF-encoding mRNAs were up-regulated in NEI cells, such as WUSCHEL-RELATED HOMEOBOX 9 (WOX9).
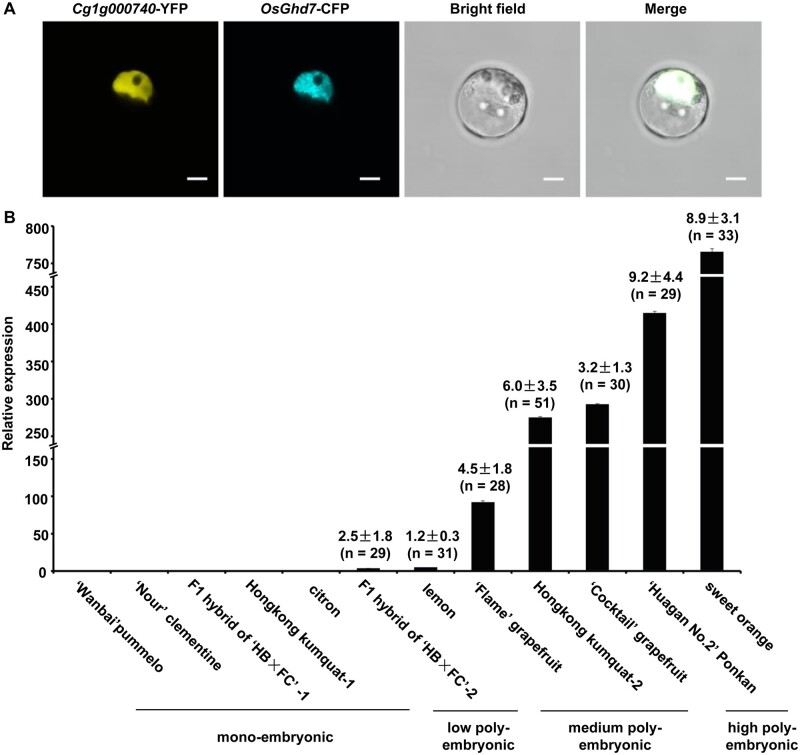
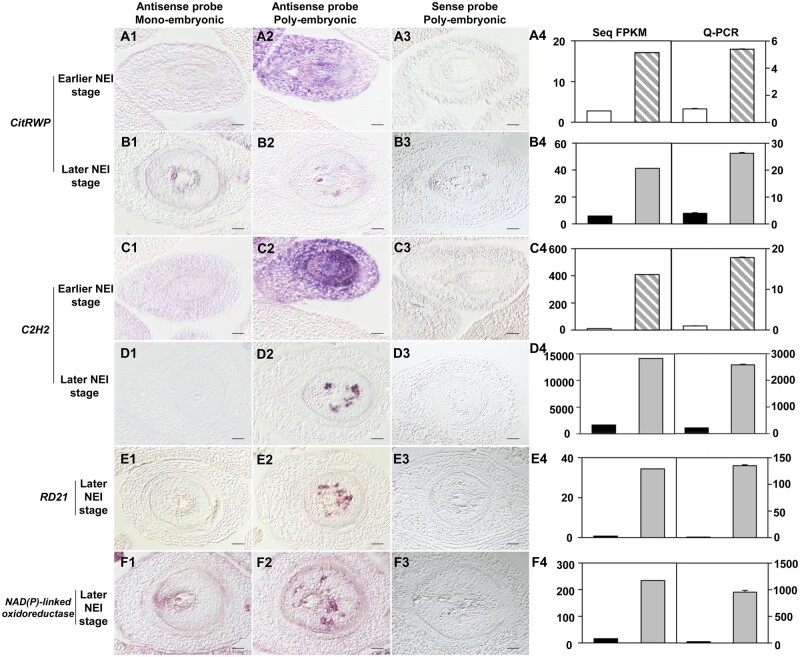
In situ hybridization experiments that utilized antisense RNA probes showed that CitRWP was expressed throughout the maternal tissue of the poly-embryonic ovule (Fig. 8A2), but was not expressed at detectable levels in the mono-embryonic ovule (Fig. 8A1) at an earlier stage of NEI cell development. At later stage of NEI cell development, CitRWP was expressed specifically in the NEI cells (Fig. 8B2), sexual embryo sac (Fig. 8B1) and juice sac in the ovary (Supplementary Fig. S11B1–B6). The expression pattern of C2H2 was similar to CitRWP. However, the expression of C2H2 was more specific to apomictic cells than the expression of CitRWP because C2H2 was not expressed in the zygotic embryo (Fig. 8C1–C3 and D1–D3) or any other tissue in the ovary (Supplementary Fig. S11C1–D6). Thus, C2H2 is a good candidate marker gene for poly-embryonic ovule and NEI cells. The fluorescence from the C2H2-YFP fusion protein colocalized with the fluorescence from rice OsGhd7-CFP (a nuclear marker) in citrus protoplasts, which demonstrates that the protein encoded by C2H2 accumulates in the nucleus (Fig. 9A). The expression level of C2H2 was quantified at an earlier stage of NEI cell development in the ovules of 12 citrus varieties using qRT-PCR. It was barely expressed in the ovules of five mono-embryonic varieties but was expressed at variable levels in the ovules of seven poly-embryonic varieties (Fig. 9B). Interestingly, the expression level of C2H2 seems positively associated with the capacity of citrus for NPE. Indeed, C2H2 was expressed at low levels in varieties with relatively low capacities for NPE (1.2–2.5 embryos per seed), at moderate levels in varieties with moderate capacities for NPE (3.2–6.0 embryos per seed) and at high levels in varieties with high capacities for NPE (8.9–9.2 embryos per seed). Two stress-responsive genes were expressed predominantly or exclusively in NEI cells. The expression of a gene encoding a cysteine protease (RD21) was up-regulated 57-fold in NEI cells (FPKM = 34) and specifically expressed in NEI cells (Fig. 8E1–E3). RD21 was not expressed in any other tissue in the ovary (Supplementary Fig. S11E1–E6), which makes it a good candidate marker gene for NEI cells. The expression of an NAD(P)-linked oxidoreductase superfamily gene was up-regulated 18-fold in NEI cells (FPKM = 234) and was specifically expressed in NEI cells, zygotic embryo (Fig. 8F1–F3) and juice sac in the ovary (Supplementary Fig. S11F1–F6). The spatiotemporal expression pattern of the four genes detected using in situ hybridization with RNA probes is consistent with the spatiotemporal expression pattern established using qRT-PCR (Fig. 8A4–F4).
Figure 9.
Expression and subcellular distribution of C2H2. (A) Subcellular localization of C2H2-YFP. Mesophyll protoplasts from citrus were co-transformed with a plasmid encoding C2H2-YFP and a plasmid encoding OsGhd7-CFP, a nuclear marker. The images were acquired with a fluorescence microscope and merged with a bright field image. Scale bars 5 µm. (B) Expression levels of C2H2 in ovules. The relative expression levels of C2H2 in ovules from 12 citrus cultivars at an earlier stage of NEI cell development were quantified using qRT-PCR. CsActin was used as the endogenous control. Data are mean values ± SD, n = 3 technical replicates from two pooled tissue samples. The cDNA template was amplified from first strand cDNA prepared from RNA that was isolated from ovules. The average number of embryos per seed was counted for each variety and is indicated above each bar.
3.6. Reactive oxygen species accumulation and oxidative stress response excitation in apomictic cells
We identified citrus homologs for seven categories of previously reported genes involved in plant reproduction: apomixis and somatic embryogenesis (SE)-associated genes (n = 79); megaspore mother cell and functional megaspore-specific genes (n = 24); meiosis-associated genes (n = 14); cell cycle-associated genes (n = 87); genes related to phytohormone biosynthesis, signalling, transport and responses (auxin, n = 82; GA, n = 21; cytokinin, n = 54); DNA methylation and small RNA pathway genes (n = 81) and oxidoreductase genes (n = 121) (Supplementary Fig. S4). Among these genes, only one category (i.e. the oxidoreductase genes) was differentially expressed in the Poly-NC relative to the Mono-NC. Among the 51 oxidoreductase genes that were expressed at earlier stage of NEI development, the expression of seven genes were up-regulated in the Poly-NC, including two genes that serve as major sources of endogenous O2– in plants (i.e. AOX1A and NDUFS4), TRX1 and four peroxidase genes. However, the expression of two peroxidase genes was down-regulated. At later stage of NEI cell development, the expression of eight genes was up-regulated in the NEI cells (i.e. AOX1A, TRX1 and five peroxidase genes), and the expression of 10 genes was down-regulated, including NDUFS4, RBOHF and eight peroxidase genes (Supplementary Fig. S4).
The O2– and H2O2 levels were detected in the poly- and mono-embryonic citrus ovules during different stages of NEI cell development using NBT and DAB staining, respectively. Although poly-embryonic ovules produced more O2– than mono-embryonic ovules at both earlier stages of NEI cell development (3 DBA) and at anthesis, O2– accumulated to barely detectable levels in both mono- and poly-embryonic ovules at 7 DAA (Supplementary Fig. S12A). The results indicate that the accumulation of O2– in ovules is important for the cell fate transition in Poly-NC but that O2– is not required after NEI cells are formed. The H2O2 content was higher in poly-embryonic than mono-embryonic ovules at both earlier and later stages of NEI cell development but lower in poly-embryonic ovules at anthesis (Supplementary Fig. S12B). Consistent with these data, response to H2O2 was enriched in both the Poly-NC and NEI cells (Fig. 3B). These results indicate that higher levels of H2O2 are important for both the cell fate transition in the Poly-NC and the identity of NEI cells. We assayed the activities of the enzymes that are responsible for the elimination of endogenous O2– and H2O2 (i.e. SODs and PODs, respectively) in ovaries. SOD activity was lower in the poly-embryonic ovaries at earlier stage of NEI cell development, consistent with the higher O2– content in poly-embryonic ovules during this stage of development (Supplementary Fig. S12C). POD activity was lower in poly-embryonic ovaries at both stages of NEI cell development, consistent with the higher H2O2 content in poly-embryonic ovules at both stages (Supplementary Fig. S12D).
4. Discussion
4.1. CitRWP and a gene encoding a C2H2 TF may initiate citrus NPE
CitRWP and C2H2 were up-regulated in apomictic cells at both stages of NEI cell development (Supplementary Fig. S10). In situ RNA hybridization revealed that CitRWP was expressed specifically in apomictic cells and the zygotic embryo during the development of NEI cells. The expression pattern of C2H2 was even more specific in that it was expressed only in apomictic cells (Fig. 8D1–D4). In Arabidopsis, the RWP-RK domain containing (RKD) TF gene RKD2 is preferentially expressed in the egg cell and embryonic cells. The ectopic expression of RKD2 led to the formation of enlarged and densely cytoplasmic embryogenic cells in ovule tissue that was reminiscent of adventitious embryony.36 In citrus, loss of CitRKD1 (i.e. CitRWP) function abolished NPE.16 We suggest that CitRWP is a marker gene for both sexual and asexual embryogenic cells in citrus and that the activated expression of CitRWP in poly-embryonic ovules activates a cell fate transition in the nucellus and specification of embryogenic cell identity. We suggest that C2H2 is a marker gene specific to apomictic cells and may work coordinately with CitRWP to initiate NPE. The correlation between increases in the gradient expression pattern of C2H2 in citrus ovules and increases in the capacity for NPE (Fig. 9B) provides evidence that C2H2 may have a dosage effect on the initiation of NPE. Whether CitRWP interacts with C2H2 and how the integration of their activities induces the initiation of NPE remains to be determined.
4.2. Reactive oxygen species may trigger the embryogenic cell fate transition and specification in apomictic cells
Previously, we and others found evidence for an oxidative stress response in the poly-embryonic ovules of citrus.15,16,18,19 Indeed, we found that the response was particularly robust in apomictic cells during the initiation of NPE (Fig. 3) and that genes encoding oxidoreductases were differentially expressed (Supplementary Fig. S4). Highly specific expression of the drought-stress inducible gene RD2137 and a salt-stress inducible gene from the NAD(P)-linked oxidoreductase superfamily38 in NEI cells makes a compelling case for the specific activation of stress response in apomictic cells (Fig. 8E2 and F2). We naturally wondered whether reactive oxygen species (ROS) accumulates in apomictic cells. Indeed, a higher level of O2– and H2O2 was detected in poly-embryonic ovules during the initiation of NPE. The signals that induce increases in the accumulation of ROS in apomictic cells and the contribution of ROS to the initiation of NPE remain open questions.
Figure 8.
Expression patterns of CitRWP, C2H2, RD21, NAD(P)-linked oxidoreductase in mono- and poly-embryonic ovules. 1–2, antisense RNA probes hybridized to the mono/poly-embryonic ovule. 3, sense RNA probes hybridized to the poly-embryonic ovule. 4, The expression level of each gene was quantified using FPKM values from RNA-Seq data (left) and qRT-PCR (right). The white, grey striped, black and grey columns represent samples from the Mono-NC, Poly-NC, SO cells and NEI cells respectively. The cDNA template was amplified from the first-strand cDNA derived from the RNA isolated from the laser-microdissected cells. Data are mean values ± SD, n = 3 technical replicates from two pooled tissue samples. Earlier and later stages of NEI cell development refer to the stages before and after the appearance of the nucellar embryo initial (NEI) cells in the nucellus. Poly/Mono-NC, poly/mono-embryonic nucellar tissue. The micrographs were prepared from longitudinal sections from single ovules at a magnification of 20×. Scale bars 40 μm.
ROS are well-known signalling molecules that regulate stem cell fate in both plants and animals.39,40 Different species of ROS play different roles in plant development. In the Arabidopsis root, a bHLH (basic helix-loop-helix) TF directly regulates the expression of a set of PODs and thus modulates the accumulation of O2– in the root meristem to maintain cell proliferation. In contrast, the accumulation of H2O2 in the elongation zone promotes cell differentiation.39 In the shoot apical meristem of Arabidopsis, accumulation of O2– in the central zone maintains stem cells. In contrast, the accumulation of H2O2 in the peripheral zone promotes cell differentiation.41 In the NPE of citrus, the accumulation of O2– and H2O2 in poly-embryonic ovules may induce a cell fate transition at an earlier stage of NEI cell development. In contrast, H2O2 may induce NEI cell specification at a later stage of NEI cell development. The up-regulated expression of particular genes that encode catalytic enzyme (i.e. AOX1A and NDUFS4), genes associated with the ‘mitochondrial respiratory chain complex I’ cellular process (i.e. major sources of endogenous O2– in plants) (Supplementary Fig. S4). Decreased SOD enzyme activity (Supplementary Fig. S12C) and down-regulated phenylpropanoid and flavonoid biosynthesis (Fig. 3A) probably promote the high level accumulation of O2– during the earlier stage of NEI cell development in poly-embryonic ovules (Supplementary Fig. S12A). Decreased POD enzyme activity (Supplementary Fig. S12D) may promote the high level accumulation of H2O2 in poly-embryonic ovules at both stages of NEI cell development (Supplementary Fig. S12B). In the poly-embryonic ovules of citrus, the NPE-controlling gene CitRWP may ultimately drive the accumulation of ROS in apomictic cells by altering gene expression patterns and ultimately, enzymes activities. The CitRWP-based mechanism that triggers the accumulation of ROS in apomictic cells remains to be elucidated.
4.3. Epigenetic regulation may induce initiation of NPE
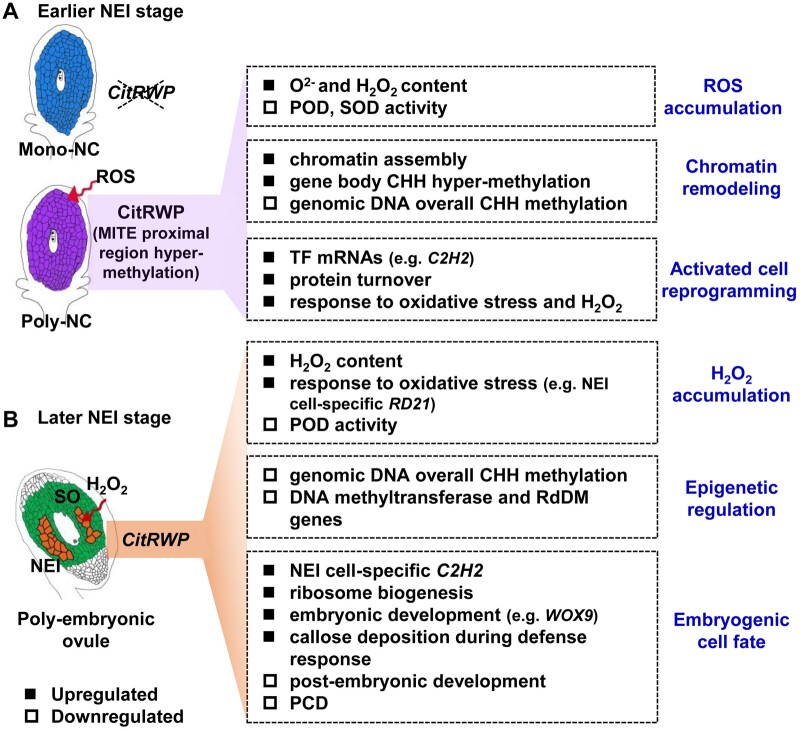
Genes associated with chromatin assembly and methylation of histone H3-K9 were enriched among the DEGs in apomictic cells (Fig. 3A and B), which indicates that a reprogramming of chromatin occurs during the initiation of NPE. The reprogramming of chromatin occurs during the somatic-to-reproductive cell fate transition in plants.42 The DNA methylation pathway is critical for distinguishing apomictic from sexual reproduction.10 Indeed, loss of methyltransferase and RdDM function down-regulates an ovule-specific chromatin-based silencing pathway and leads to apomictic reproduction.10–12 In citrus apomictic cells, global CHH methylation levels were decreased (Fig. 5). In NEI cells, the CHH hypo-methylation of DNA was prominent, especially in retrotransposons (Fig. 6B). The expression of two DNA methyltransferase genes and four RdDM pathway genes was down-regulated in NEI cells (Supplementary Fig. S4). Among these genes, DECREASE IN DNA METHYLATION 1 (DDM1) is involved in the maintenance of methylation at both CG and non-CG sites in Arabidopsis. A mutation in DDM1 induces a profound loss of methylation for particular TEs and leads to strong transcriptional activation of TEs.43NRPE5 is the fifth largest subunit of Pol V. This special subunit mediates RdDM.44 The lower level of CHH methylation in the transposons of NEI cells might be caused by the down-regulated expression of genes associated with DNA methylation. In Arabidopsis and maize, GA-like phenotypes were observed in the loss-of-function mutants that are deficient in DNA methylation, especially in mutants deficient in RdDM (e.g. dmt, ago9, sgs3 and nrpd1a nrpd1b, an RNA Polymerase IV and V double mutant).10–12 In our study, the global CHH hypo-methylation and down-regulated expression of DNA methyltransferase and RdDM pathway genes in apomictic cells indicates that the regulation of RdDM might also contribute for SA. Collectively, we suggest that the reprogramming of chromatin, especially the CHH hypo-methylation of DNA that is associated with the down-regulated expression of genes that encode DNA methyltransferases and components of the RdDM pathway (Supplementary Fig. S4), may change the epigenetic and transcriptional status of apomictic cells and thus may facilitate cell fate transitions towards embryogenesis during the initiation of NPE in citrus (Fig. 10).
Figure 10.
Working model for the initiation of nucellar poly-embryogenesis in citrus. Earlier (A) and later (B) stages of NEI cell development. The solid/hollow quadrangles indicate genes, metabolites, biological processes and epigenetic modifications that are either up- or down-regulated in apomictic cells. The earlier and later stages of NEI cell development refer to the stages before and after the appearance of nucellar embryo initial (NEI) cells in the nucellus. Poly/Mono-NC, poly/mono-embryonic nucellar tissue.
On the other hand, the methylation of PCGs is positively associated with the expression levels of genes that encode pathways that were enriched during NPE and with the expression levels of genes that are required for NPE in citrus, such as genes that are up-regulated in the Poly-NC (Fig. 7A), ribosome pathway (Supplementary Fig. S6A), plant hormone signal transduction pathway (Supplementary Fig. S6D), UBQ6 and CitRWP (Fig. 7B and C). A growing number of genome-wide methylation studies provides evidence that the relationship between DNA methylation and gene expression is complex.45 In most angiosperms, genes with gene body methylation are generally constitutively expressed.46–48 Although DNA methylation in the promoter region usually inhibits gene transcription, it promotes gene transcription in some cases (e.g. ROS1 from Arabidopsis and genes that inhibit fruit ripening in tomato).46,49–52 In the case of CitRWP, hyper-methylation of the promoter region containing a MITE insertion might activate gene transcription in the poly-embryonic ovules by enhancing the binding of particular transcription activators or by inhibiting the binding of particular transcription repressors. Together, we suggest that the global reduction of CHH methylation levels in the DNA of apomictic cells may derepress retrotransposons and thus alter the expression of adjacent genes that regulate development. The methylation of PCGs may promote the expression of critical biological processes, pathways and key genes that induce the initiation of NPE. ROS is an attractive candidate for the signal that triggers the epigenetic reprogramming of apomictic cells in citrus. ROS affects epigenetic regulatory systems, global DNA methylation and the regulation of oxidative stress. Indeed, ROS have been proposed to regulate sex/apomixis switching in apomictic plants.53,54 The stress induced accumulation of ROS and the epigenetic reorganization of plant cells was also reported to promote in vitro SE.55 Therefore, we suggest that the accumulation of ROS may trigger the cell fate transition and SE in citrus ovules by inducing the epigenetic reprogramming of apomictic cells.
4.4. Citrus NPE initiation shares common features with GA and SE
To identify the important biological processes, pathways and key genes that are activated and repressed by epigenetic reprogramming during the initiation of NEI cells and to specify the identity of apomictic cells in citrus, we investigated the enriched GO terms shared among different types of apomixis and SE. In the apomictic cells of citrus, protein turnover was the most significantly enriched processes (Fig. 3; Supplementary Figs. S2B and S6A). Protein turnover was also dramatically induced in the pre-meiotic germinal cells of Arabidopsis56 and in the AI cells of Hieracium and Boechera—two GA plants.7,57 Together, these data provide evidence that protein turnover is activated in both sexual reproductive cells and AI cells and that a ribosome-based regulatory mechanism may generally contribute to apomixis in plants. In the NEI cells of citrus, the expression of genes associated with auxin polar transport process and auxin biosynthesis, signalling and transport pathway genes was down-regulated. In apomictic Hieracium, earlier and more AI-like cells appeared in the ovule when polar auxin transport was inhibited, and changes in the frequency and location of AI cells were observed when the sensitivity to auxin was increased.58 Therefore, inhibition of polar auxin transport and auxin responses might promote the initiation of both gametophytic and SA and help to specify embryogenic cell fate.
In NEI cells, activation of callose deposition may lead to callose deposition in the cell wall, which explains the thickened cell wall of NEI cells. Callose deposition at the plasmodesmata is required for the symplasmic isolation of embryogenic cells from non-embryogenic cells during SE and the establishment of cell totipotency.59 We suggest that callose deposition may serve to isolate the NEI cells from the surrounding nucellar SO cells and thus, to help maintain embryogenic cell fate and to facilitate the subsequent SE. Callose accumulates in the walls of cells undergoing megasporogenesis and SE but not in GA initial cells.60 This indicates that NEI cells share identity with embryogenic cells instead of GA initial cells. In addition, genes associated with embryonic development process were enriched among the up-regulated genes in citrus NEI cells (Fig. 3B). In contrast, genes associated with gametophyte development process were enriched among the up-regulated genes in GA initial cells from Hieracium.7 These data reflect the distinct embryogenic cell fates of NEI cells and GA initial cells. Notably, genes associated with PCD process were enriched among the down-regulated genes in NEI cells (Fig. 3). In addition, the stress-responsive RD21 was specifically expressed in citrus NEI cells (Fig. 8). The homolog of RD21 in Arabidopsis acts as a negative regulator of sphingolipid-induced cell death.61 Therefore, we suggest that the specific and abundant expression of RD21 and the down-regulation of genes associated with PCD process may have protected the NEI cells from degradation alongside the surrounding nucellar SO cells after anthesis. Thus, the NEI cells can divide and form the embryonic cell mass that enters the enlarging embryo sac and develop into asexual embryos alongside the zygotic embryo.
Based on the transcriptome and DNA methylome profiles in apomictic cells, we suggest a working model for the initiation of NPE in citrus (Fig. 10). In our model, the expression of the key NPE-controlling gene CitRWP is activated in the poly-embryonic citrus ovule, which is associated with the hyper-methylation of DNA in the NPE-specific MITE insertion region in the promoter of CitRWP. The transcriptional upregulation of CitRWP results in the epigenetic regulation and ROS accumulation in apomictic cells. These effects activate a cell fate transition in the poly-embryonic nucellus and specify embryogenic cell identity in NEI cells. NPE in citrus shares common features with GA during the initiation of apomixis, in that the somatic cells undergo reprogramming but later diverge to SE, when NEI cells have acquired an embryogenic cell fate.
Supplementary Material
Acknowledgements
We thank Professor Robert B. Goldberg and his laboratory members at the University of California, Los Angeles, especially Min Chen, for instruction in laser microdissection and our colleagues Prof. Fang Yang and her laboratory for technical instructions in RNA in situ hybridization and Prof. Robert M. Larkin for critical reading of the manuscript. This research was financially supported by the Ministry of Science and Technology of China (no. 2018YFD1000106), the National Natural Science Foundation of China (nos. 31872051, 31630065), the Fundamental Research Funds for Central Universities (no. 2662018PY013) and Hubei Hongshan Laboratory.
Accession numbers
The data that support the findings of this study are openly available in NCBI database under the BioProject accession PRJNA706142, PRJNA706473.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Brukhin V. 2017, Molecular and genetic regulation of apomixis, Russ. J. Genet., 53, 943–64. [Google Scholar]
- 2. Conner J.A., Mookkan M., Huo H.Q., Chae K., Ozias-Akins P.. 2015, A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant, Proc. Natl. Acad. Sci. USA., 112, 11205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khanday I., Skinner D., Yang B., Mercier R., Sundaresan V.. 2019, A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds, Nature, 565, 91–5. [DOI] [PubMed] [Google Scholar]
- 4. Wang C., Liu Q., Shen Y., et al. 2019, Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes, Nat. Biotechnol., 37, 283–6. [DOI] [PubMed] [Google Scholar]
- 5. Sharbel T.F., Voigt M.L., Corral J.M., et al. 2010, Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns, Plant Cell, 22, 655–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mau M., Corral J.M., Vogel H., et al. 2013, The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species, Plant Physiol., 163, 1640–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okada T., Hu Y., Tucker M.R., et al. 2013, Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis, Plant Physiol., 163, 216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt A., Schmid M.W., Klostermeier U.C., et al. 2014, Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation, PLoS Genet., 10, e1004476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Juranic M., Tucker M.R., Schultz C.J., et al. 2018, Asexual female gametogenesis involves contact with a sexually-fated megaspore in apomictic Hieracium, Plant Physiol., 177, 1027–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Aguilar M., Michaud C., Leblanc O., Grimanelli D.. 2010, Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes, Plant Cell., 22, 3249–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., et al. 2010, Control of female gamete formation by a small RNA pathway in Arabidopsis, Nature, 464, 628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grimanelli D. 2012, Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms, Curr. Opin. Plant Biol., 15, 57–62. [DOI] [PubMed] [Google Scholar]
- 13. Nakano M., Shimada T., Endo T., et al. 2012, Characterization of genomic sequence showing strong association with polyembryony among diverse Citrus species and cultivars, and its synteny with Vitis and Populus, Plant Sci., 183, 131–42. [DOI] [PubMed] [Google Scholar]
- 14. Kepiro J.L., Roose M.L.. 2010, AFLP markers closely linked to a major gene essential for nucellar embryony (apomixis) in Citrus maxima × Poncirus trifoliata, Tree Genet. Genomes, 6, 1–11. [Google Scholar]
- 15. Wang X., Xu Y.T., Zhang S.Q., et al. 2017, Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction, Nat. Genet., 49, 765–72. [DOI] [PubMed] [Google Scholar]
- 16. Shimada T., Endo T., Fujii H., et al. 2018, MITE insertion-dependent expression of CitRKD1 with a RWP-RK domain regulates somatic embryogenesis in citrus nucellar tissues, BMC Plant Biol., 18, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakano M., Kigoshi K., Shimizu T., et al. 2013, Characterization of genes associated with polyembryony and in vitro somatic embryogenesis in Citrus, Tree Genet. Genomes, 9, 795–803. [Google Scholar]
- 18. Kumar V., Malik S.K., Pal D., Srinivasan R., Bhat S.R.. 2014, Comparative transcriptome analysis of ovules reveals stress related genes associated with nucellar polyembryony in citrus, Tree Genet. Genomes, 10, 449–64. [Google Scholar]
- 19. Long J.M., Liu Z., Wu X.M., et al. 2016, Genome-scale mRNA and small RNA transcriptomic insights into initiation of citrus apomixis, J. Exp. Bot., 67, 5743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen M., Lin J.Y., Wu X.M., et al. 2021, Comparative analysis of embryo proper and suspensor transcriptomes in plant embryos with different morphologies, Proc. Natl. Acad. Sci. USA., 118, e2024704118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu G.A., Prochnik S., Jenkins J., et al. 2014, Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication, Nat. Biotechnol., 32, 656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mihaela P., Daehwan K., Geo M.P., Jeffrey T.L., Steven L.S.. 2016, Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown, Nat. Protoc., 11, 1650–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson M.D., McCarthy D.J., Smyth G.K.. 2010, edgeR: a bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du Z., Zhou X., Ling Y., Zhang Z., Su Z.. 2010, agriGO: a GO analysis toolkit for the agricultural community, Nucleic Acids Res., 38, W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krueger F., Andrews S.R.. 2011, Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications, Bioinformatics, 27, 1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang S., Yin Z.P., Wu X.M., et al. 2020, Assembly of Satsuma mandarin mitochondrial genome and identification of cytoplasmic male sterility–specific ORFs in a somatic cybrid of pummelo, Tree Genet. Genomes, 16, 84. [Google Scholar]
- 27. Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E.. 2013, Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome, Cell, 152, 352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruntman E., Qi Y.J., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R.. 2008, Kismeth: analyzer of plant methylation states through bisulfite sequencing, BMC Bioinformatics, 9, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R., Fang Y.N., Wu X.M., et al. 2020, The miR399-CsUBC24 module regulates reproductive development and male fertility in Citrus, Plant Physiol, 183, 1681–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang F., Bui H.T., Pautler M., et al. 2015, A maize glutaredoxin gene, Abphyl2, regulates shoot meristem size and phyllotaxy, Plant Cell, 27, 121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xue W., Xing Y., Weng X., et al. 2008, Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice, Nat. Genet., 40, 761–7. [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Sun P.P., Chen C.L., Wang Y., Fu X.Z., Liu J.H.. 2011, An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis, J. Exp. Bot., 62, 2899–914. [DOI] [PubMed] [Google Scholar]
- 33. Gao S., Yan R., Cao M., Yang W., Wang S., Chen F.. 2008, Effects of copper on growth, antioxidant enzymes and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedling, Plant. Soil Environ., 54, 117–22. [Google Scholar]
- 34. Enke R.A., Dong Z.C., Bender J.. 2011, Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana, PLoS Genet., 7, e1002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Y.T., Jia H.H., Wu X.M., Koltunow A.M., Deng X.X., Xu Q.. 2021, Regulation of nucellar embryony, a mode of sporophytic apomixis in Citrus resembling somatic embryogenesis, Curr. Opin. Plant Biol., 59, 101984. [DOI] [PubMed] [Google Scholar]
- 36. Koszegi D., Johnston A.J., Rutten T., et al. 2011, Members of the RKD transcription factor family induce an egg cell-like gene expression program, Plant J., 67, 280–91. [DOI] [PubMed] [Google Scholar]
- 37. Liu Y., Wang K., Cheng Q., et al. 2020, Cysteine protease RD21A regulated by E3 ligase SINAT4 is required for drought-induced resistance to Pseudomonas syringae in Arabidopsis, J. Exp. Bot., 71, 5562–76. [DOI] [PubMed] [Google Scholar]
- 38. Liu J.X., Srivastava R., Che P., Howell S.H.. 2007, Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling, Plant J., 51, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsukagoshi H., Busch W., Benfey P.N.. 2010, Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root, Cell, 143, 606–16. [DOI] [PubMed] [Google Scholar]
- 40. Zhou G., Meng S., Li Y., Ghebre Y.T., Cooke J.P.. 2016, Optimal ROS signaling is critical for nuclear reprogramming, Cell Rep., 15, 919–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeng J., Dong Z., Wu H., Tian Z., Zhao Z.. 2017, Redox regulation of plant stem cell fate, Embo J., 36, 2844–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. She W., Grimanelli D., Rutowicz K., et al. 2013, Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants, Development, 140, 4008–19. [DOI] [PubMed] [Google Scholar]
- 43. Zemach A., Kim M.Y., Hsieh P.H., et al. 2013, The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin, Cell., 153, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ream T.S., Haag J.R., Wierzbicki A.T., et al. 2009, Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II, Mol. Cell, 33, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H.M., Lang Z.B., Zhu J.K.. 2018, Dynamics and function of DNA methylation in plants, Nat. Rev. Mol. Cell Biol., 19, 489–506. [DOI] [PubMed] [Google Scholar]
- 46. Zhang X., Yazaki J., Sundaresan A., et al. 2006, Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis, Cell, 126, 1189–201. [DOI] [PubMed] [Google Scholar]
- 47. Takuno S., Gaut B.S.. 2013, Gene body methylation is conserved between plant orthologs and is of evolutionary consequence, Proc. Natl. Acad. Sci. USA., 110, 1797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Niederhuth C.E., Bewick A.J., Ji L.X., et al. 2016, Widespread natural variation of DNA methylation within angiosperms, Genome Biol., 17, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang H.M., Zhu J.K.. 2012, Active DNA demethylation in plants and animals, Cold Spring Harb. Symp. Quant. Biol., 77, 161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lei M.G., Zhang H.M., Julian R., Tang K., Xie S.J., Zhu J.K.. 2015, Regulatory link between DNA methylation and active demethylation in Arabidopsis, Proc. Natl. Acad. Sci. USA., 112, 3553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williams B.P., Pignatta D., Henikoff S., Gehring M.. 2015, Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat, PLoS Genet., 11, e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lang Z.B., Wang Y.H., Tang K., et al. 2017, Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit, Proc. Natl. Acad. Sci. USA., 114, E4511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Albertini E., Barcaccia G., Carman J.G., Pupilli F.. 2019, Did apomixis evolve from sex or was it the other way around?, J. Exp. Bot., 70, 2951–64. [DOI] [PubMed] [Google Scholar]
- 54. Schmidt A. 2020, Controlling apomixis: shared features and distinct characteristics of gene regulation, Genes, 11, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feher A. 2015, Somatic embryogenesis – stress-induced remodeling of plant cell fate, Biochim. Biophys. Acta., 1849, 385–402. [DOI] [PubMed] [Google Scholar]
- 56. Yuan T.L., Huang W.J., He J., Zhang D., Tang W.H.. 2018, Stage-specific gene profiling of germinal cells helps delineate the mitosis/meiosis transition, Plant Physiol., 176, 1610–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zuhl L., Volkert C., Ibberson D., Schmidt A.. 2019, Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera, J. Exp. Bot., 70, 5643–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tucker M.R., Okada T., Johnson S.D., Takaiwa F., Koltunow A.M.. 2012, Sporophytic ovule tissues modulate the initiation and progression of apomixis in Hieracium, J. Exp. Bot., 63, 3229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Godel-Jedrychowska K., Kulinska-Lukaszek K., Horstman A., et al. 2020, Symplasmic isolation marks cell fate changes during somatic embryogenesis, J. Exp. Bot., 71, 2612–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tucker M.R., Paech N.A., Willemse M.T.M., Koltunow A.M.G.. 2001, Dynamics of callose deposition and β-1,3-glucanase expression during reproductive events in sexual and apomictic Hieracium, Planta., 212, 487–98. [DOI] [PubMed] [Google Scholar]
- 61. Ormancey M., Thuleau P., van der Hoorn R.A.L., et al. 2019, Sphingolipid-induced cell death in Arabidopsis is negatively regulated by the papain-like cysteine protease RD21, Plant Sci., 280, 12–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.