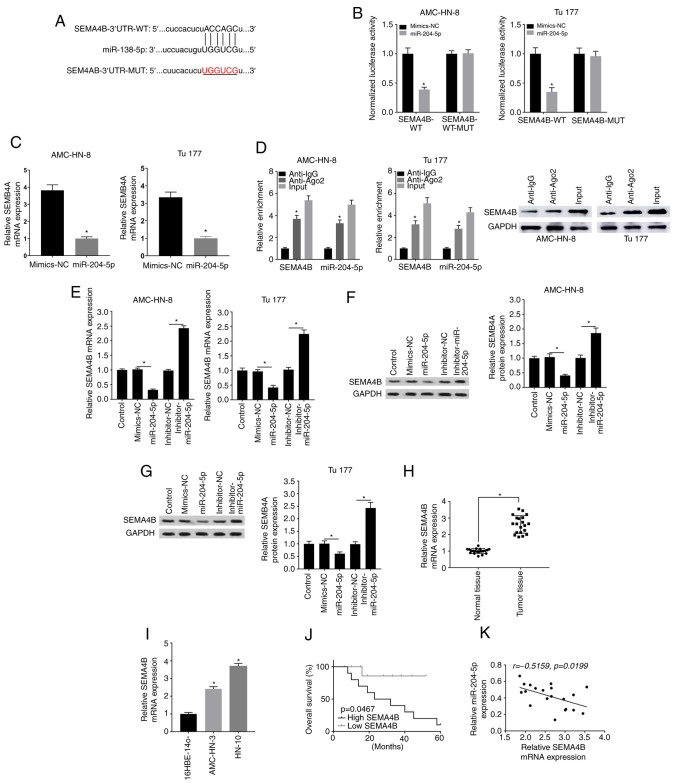
Figure 6.
SEMA4B is targeted by miR-204-5p in LSCC. (A) Target complementary sequence between miR-204-5p and SEMA4B was analyzed using bioinformatics analysis. (B) Targeting association between SEMA4B and miR-204-5p was verified using dual luciferase reporter assay. (C) Effect of overexpressed miR-204-5p on SEMA4B expression was analyzed using RT-qPCR. *P<0.05 vs. mimics-NC. (D) Enrichment of SEMA4B or miR-204-5p was measured by RNA immunoprecipitation assay in AMC-HN-3 and Tu177 cells incubated with Ago2 or IgG. *P<0.05 vs. anti-IgG. (E) Effect of inhibitor miR-204-5p on SEMA4B expression was measured using RT-qPCR. Effect of inhibitor miR-204-5p on SEMA4B expression was evaluated using western blotting in (F) AMC-HN-3 and (G) Tu177 cells. (H) Relative SEMA4B expression in LSCC and normal tissues was detected using RT-qPCR. *P<0.05. (I) Relative SEMA4B expression in LCSS cell lines and 16HBE-14o cells was detected using RT-qPCR. *P<0.05 vs. 16HBE-14o cells. (J) Overall survival rate according to SEMA4B expression was analyzed using Kaplan-Meier survival analysis. (K) Correlation analysis between SEMA4B and miR-204-5p expression. The data are presented as the mean ± SD of three independent experiments. SEMA4B, class 4B semaphorins; RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; NC, negative control; Ago2, argonaute 2; WT, wild-type; MUT, mutant; LSCC, laryngeal squamous cell carcinoma.