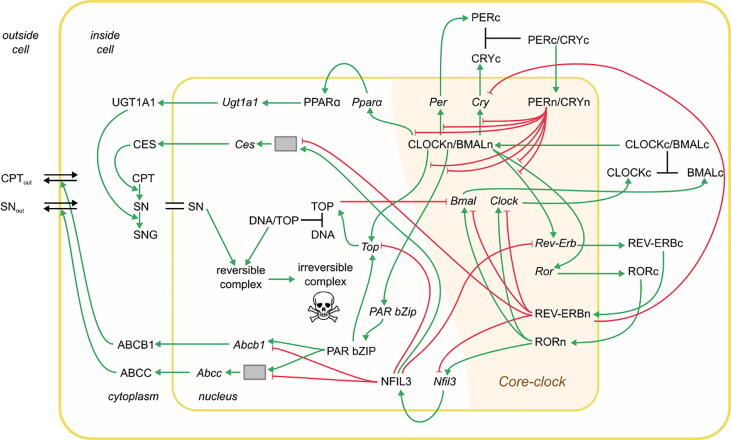
Fig. 4.
Model of the interplay between irinotecan PK-PD and the core clock. Irinotecan treatment is simulated by a transcriptional-translational network that comprises the core clock, irinotecan-relevant genes, and the PK-PD of irinotecan. Different types of interactions are represented among the elements of the network: inhibition (red arrows with flat arrowhead) and activation interactions (green arrows); complex formation (black lines). Grey boxes represent post-transcriptional sub-networks necessary for a model fit to the data. The double black line indicates equal concentrations between nucleus and cytoplasm, the double black line with arrowheads indicates CPT-11 and SN-38 cellular transport inside and outside of the cell. All indicated molecular interactions are based on experimental evidence from a number of different sources and corresponding references are provided in Supplementary Table 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)