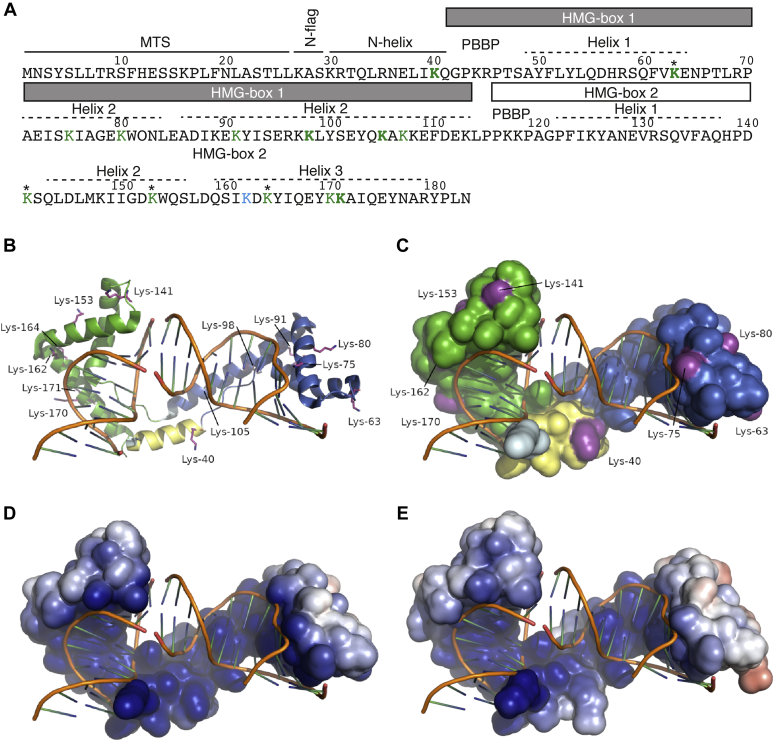
Figure 4.
An overview of a single monomer bound to DNA from the Abf2p complex structure and a model of its succinylated form. A, sequence of Abf2p with highlighted structural regions based on the 3D structure of the protein (58). Lysines identified in a succinylated form in vivo in this study are in green. K162 identified by Weinert et al. (19) is in blue. The lysine residues having in vitro modified/unmodified ratios of more than 0.25 are in bold (panel E). MS analysis of Abf2p succinylated in vitro identified peptides containing Ksucc corresponding to all lysine residues except K27, K30, and K45. Lysine residues marked with asterisks (∗) were also shown to be acetylated in vivo (46). B, an overview of half the Abf2p–DNA complex. The lysines found to be succinylated in this study are colored magenta, shown as sticks, and labeled. HMG box 1 is colored blue, HMG box 2 is green, the N-helix is yellow, and the N-flag is light cyan. This same coloring is used in the next panel. C, the same complex and the same view as before but showing the solvent-accessible surface area. Not all the succinylated lysines are visible. D, the solvent-accessible surface of Abf2p colored by electrostatic surface potential (the range is ±5 kT/e). E, the solvent-accessible surface of succinylated Abf2p colored by electrostatic surface potential (the same range as before). For the succinylated form, five residues were selected that had the greatest ratio of modified to unmodified forms in vitro and that were also found to be succinylated in vivo. B, basic amino acid; HMG box, high-mobility group box; MTS, mitochondrial targeting sequence.