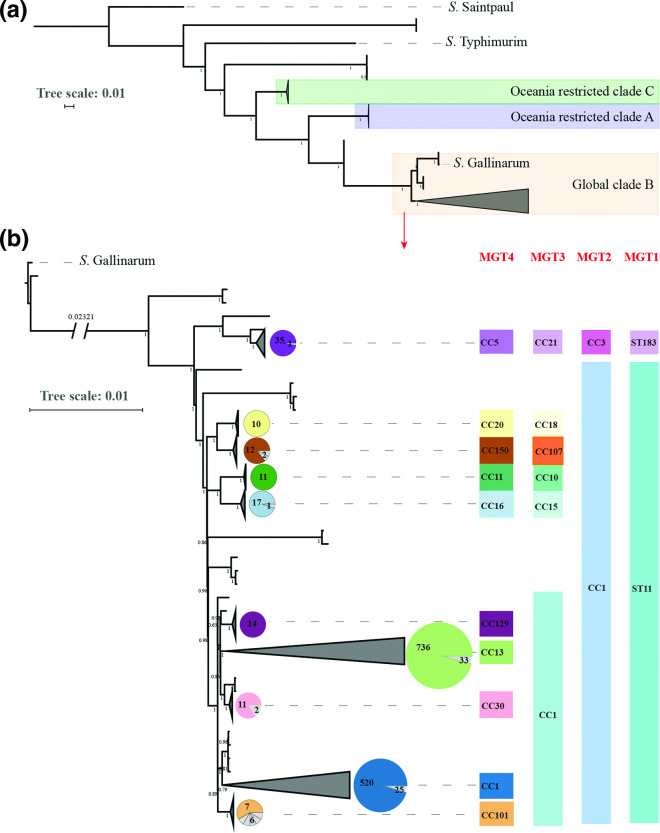
Fig. 6.
Global phylogenetic structure of S. Enteritidis. A total of 1508 genomes were randomly sampled from each ST in MGT6. A phylogenetic tree was constructed by ParSNP v1.2, which called core-genome SNPs extracted from alleles. The tree scales indicated 0.01 substitutions per locus. The number on internal branches represented the percentage of bootstrap support. (a) Three clades were identified which were concordant with previous findings: The predominant clade B, highlighted in yellow, is the global epidemic clade which was phylogenetically closer to S. Gallinarum than clade A and C in purple and green background which are limited in Oceania [11]. (b) Clade B was expanded into a more detailed phylogeny with S. Gallinarium as an outgroup. Ten main lineages were defined where each lineage had more than ten isolates. STs of the seven gene MLST (or MGT1) and CCs from MGT2 to MGT4 are shown in different colours for each of the 10 lineages. The pie charts for each lineage represent the proportion of the isolates belonged to the same MGT4-CC shown on the right side.