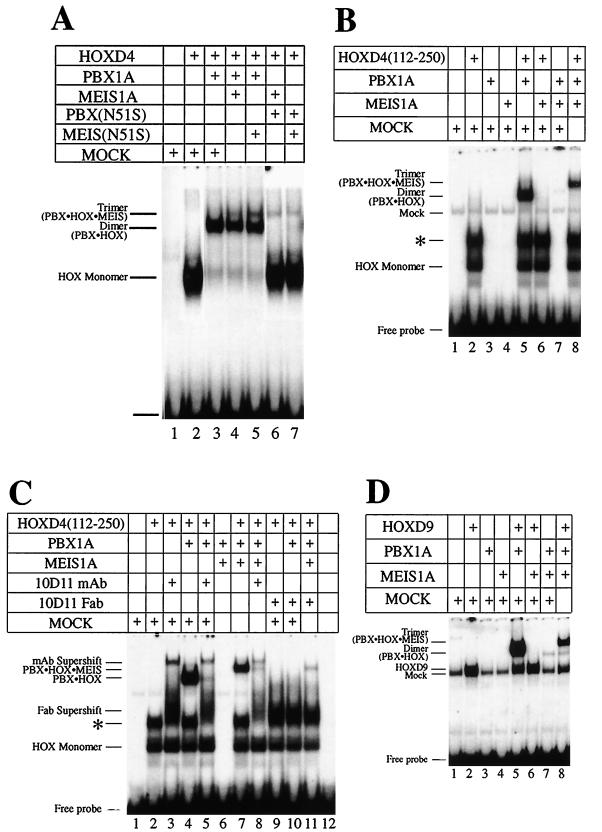
FIG. 4.
Formation of a trimeric complex by MEIS(N51S). (A) MEIS1A is the non-DNA-binding component in the trimer with PBX-HOXD4 as seen in EMSA. Full-length HOXD4 binds an A6 site (Fig. 1A) as a monomer (lane 2) and as a cooperative dimer with PBX (lane 3). Addition of MEIS1A or MEIS(N51S) leads to the formation of an additional low-mobility complex (lanes 4 and 5). Loss of the cooperative dimer and retention of the presumptive trimer were observed when PBX(N51S) was used in the presence of MEIS1A and HOXD4 or of MEIS(N51S) and HOXD4 (lanes 6 and 7). (B) Deletion of aa 1 to 111 from HOXD4 reveals a negative effect of this region on the formation of the trimer. Deleting the first 111 aa resulted in a more robust trimeric complex: all of the heterodimer is converted to presumptive heterotrimer in lane 8 (compare to lane 4 in Fig. 4A). Also compare lanes 4 and 7 in Fig. 1C. (C) A MAb against the HOXD4 YPWM prevents the formation of the trimeric complex in EMSA. Addition of MAb 10D11 (54) or the respective Fab fragments led to the loss of the dimeric and trimeric complexes (compare lane 5 to lane 4, lane 8 to lane 7, and lanes 10 and 11 to lanes 4 and 7). (D) MEIS1A can be a non-DNA-binding component in a trimer with PBX1A-HOXD9. Addition of MEIS1A to PBX1A and HOXD9 led to the formation of a low-mobility band in EMSA (compare lane 8 to lane 5). On the site used here, no or little DNA-binding activity was observed for MEIS1A as a monomer (lane 4) or in combination with HOXD9 (lane 6) or PBX1A (lane 7), suggesting that DNA binding by MEIS1A is not involved in the formation of the presumptive trimer. ∗, an apparent dimer of HOXD4 that we have shown to be very unstable (data not shown). A 32P-labeled T6 target site (Fig. 1A) was used as a probe.