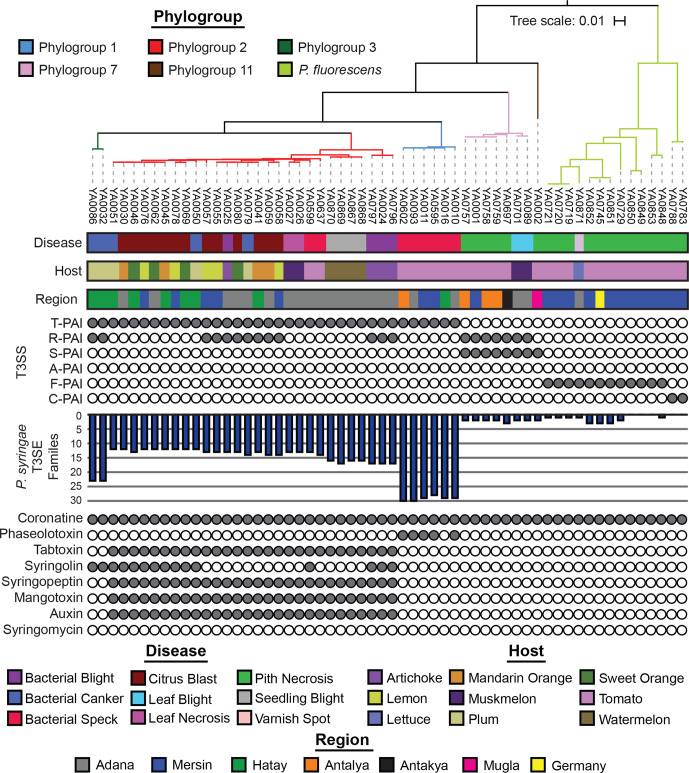
Fig. 4.
T3SS, T3SE and phytotoxin repertoires for each of the 58 representative Pseudomonas strains that were whole-genome sequenced. The phylogenetic tree was generated from a concatenated core-genome amino acid alignment using FastTree, with an SH-Test branch support cut-off of 50 %. The tree was rooted at the base of the P. syringae species complex and the tree scale reflects the number of stubstitutions per site. The six T3SSs analysed include the T-PAI from P. syringae PtoDC3000, the R-PAI from P. syringae Pph1448a, the S-PAI from P. syringae PchICMP3353, the A-PAI from P. syringae PcoICMP19117, the F-PAI from P. fluorescens PgeBBc6R8 and the C-PAI from P. corrugata PcoF113. The presence–absence of 70 established P. syringae T3SEs in each genome was used to quantify the collective effector repertoires in each strain. A phytotoxin was considered present if more than 50 % of the known protein sequences involved in the synthesis pathway had significant tblastn hits (1×10−5, >80 % identity) in the genome.