Abstract
Hexavalent Chromium [Cr (VI)] is an established toxicant, carcinogen, and a significant source of public health concern. The multicopy ribosomal DNA (rDNA) array has been mechanistically implicated in aging and cancer, is the most evolutionarily conserved segment of the human genome, and gives origin to nucleolus, a nuclear organelle where ribosomes are assembled. Here we show that exposure to Cr (VI) induces instability in the rDNA, triggering cycles of rapid, specific, and transient amplification and contraction of the array in human cells. The dynamics of environmentally responsive rDNA copy number (CN) amplification and contraction occurs at doses to which millions of individuals are regularly exposed. Finally, analyses of human populations occupationally exposed to Cr (VI) indicate that environmental exposure history and drinking habits but not age shape extensive naturally occurring rDNA copy number variation. Our observations identify a novel pathway of response to hexavalent chromium exposure and raise the prospect that a suite of environmental determinants of rDNA copy number remain to be discovered.
Hexavalent Chromium [Cr (VI)] is an established human toxicant, carcinogen, and a significant source of public health concern. Human exposure to Cr (VI) occurs from both natural and anthropogenic sources with millions of exposed individuals worldwide, including occupationally exposed workers (Bianchi et al. 1980; Cohen et al. 1993; Tully et al. 2000; Bagchi et al. 2002; Chen and Shi 2002; Salnikow and Zhitkovich 2008; Arita and Costa 2009). Over 2,500 tons of Chromium is emitted to the atmosphere annually from anthropogenic sources in the US alone. Occupational exposure can be dramatic in electroplating factories and welding environments, but exposure through the drinking water is also a perennial source of concern (Langard 1990; Cohen et al. 1993; Gibb et al. 2000). Chromium content in the general population was estimated at 0.006 μg/L (0.01–0.17 μg/L) in serum while occupational exposed workers in the US and England can display substantially higher levels (e.g., up to 216 μg/L in blood of workers from pigment factories). Cr (VI) undergoes intracellular reduction to Cr (III) by reductive agents; the reactive species produced during the reduction of Cr (VI) causes cytotoxicity and genetic damage (Shi et al. 1999; Liu and Shi 2001; Costa and Klein 2006). Multiple cellular pathways can repair Cr-damaged DNA: DNA double-strand breaks repair (DSB), mismatch repair (MMR), transcription-coupled nucleotide excision repair (TCR), among others (Hartwig et al. 2002; Hartwig and Schwerdtle 2002; Reynolds et al. 2004; Wise et al. 2008). One mechanism of chromium induced DNA damage is through the formation of Cr-DNA adducts initiated by Cr (III) that preferentially occur at GG di-nucleotides (Arakawa et al. 2012) and inhibit DNA replication and RNA transcription.
The ribosomal DNA (rDNA) array is an essential GC-rich multicopy DNA element of eukaryotic genomes. The 45S rDNA repeat unit is comprised of the 18S, 5.8S and 28S rDNAs; the rDNA array gives origin to the nucleolus, an energy intensive nuclear organelle and site of ribosomal RNA (rRNA) synthesis. Classical estimates of rDNA copy number (rDNA CN) indicated that Drosophila, mice, and humans harbored ~300 rDNA copies (Henderson et al. 1972; Henderson et al. 1974; Long and Dawid 1980; Stage and Eickbush 2007; Stults et al. 2008). However, seminal population studies also documented extensive naturally occurring rDNA CN variation (Lyckegaard and Clark 1989; Rustchenko et al. 1993; Stults et al. 2008), which has been confirmed with sequencing technologies (Gibbons et al. 2014; Gibbons et al. 2015). Substantial changes in rDNA CN were observed even between parents and offspring (Stults et al. 2008). This included the regular detection of completely new rDNA CN alleles in the offspring that were not present in either parent (Stults et al. 2008). Higher rDNA CN in blood has been recently suggested as a risk factor for increased lung cancer risk (Hosgood et al. 2019).
Nucleoli are dynamic structures that vary, both in size and appearance, from one cell type to another, and play a central role in myriad cellular processes (Pederson 1998; McStay and Grummt 2008; Boulon et al. 2010; Mayer and Grummt 2014). Nucleoli are the site of assembly of the ribosome, an essential cellular machine that is comprised of more than 80 proteins and 4 ribosomal RNAs (the three 45S rRNAs - 18S, 5.8S, and 28S rRNA – and the 5S rRNA). Over 60–90% of all cellular RNAs are rRNAs, which are synthesized at exceptionally high rates to supply the thousands of ribosomes produced per minute in eukaryotic cells. Variation in the rDNA CN among individuals is functionally significant, with hundreds of genes across the genome displaying naturally occurring gene expression variation that was significantly associated with variation in rDNA CN (Gibbons et al. 2014). Impacts of chromium exposure on the rDNA arrays have not been reported.
Here we show that the human rDNA CN is exquisitely responsive to Cr (VI). Treatment at concentrations to which millions of individuals are regularly exposed triggered the rapid but transient amplification of the rDNA approximately 48hs after the onset of exposure in both immortal cell lines and primary cell cultures. Moreover, analysis of a population of workers chronically exposed to Cr (VI) showed significantly lower rDNA CN in these individuals and a positive association with drinking habits in both exposed and non-exposed populations, whereas individual age was not associated with rDNA CN. Collectively, the data point to a model of genetic variation in the rDNA that is exquisitely dynamic and environmentally responsive.
Methods
Cell cultures and treatments
The human lymphoblastoid cell line (LCL) NA12043 was obtained from the Coriell Cell Repository. The cells were cultured in T25 flasks in RPMI 1640 (GIBCO™) medium supplemented with 10% fetal bovine serum (FBS) in a humidified incubator with 5% CO2 at 37°C. Hela cells were cultured in T200 flasks with DMEM (Doubecco’s modified Eagle’s medium, GIBCO™) medium supplemented with 10% FBS and 1% glutamine in ambient condition as described above. Chemicals [5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), RNA Polymerase III Inhibitor 1, 5-Aza-cytidine, 5-Aza-2’-deoxycytidine, Cisplatin, and Actinomycin D (AMD)] were dissolved in dimethyl sulphoxide (DMSO). Maximal final concentration of DMSO used in cell culture was kept below 0.1%. Potassium dichromate (K2Cr2O7) was prepared in RPMI-1640 without serum. The exposure window for all treatments (K2Cr2O7, CdCl2 and cellular inhibitors below) was 24h: cells were exposed for 24h, at which point the media was removed and cells washed twice with PBS, followed by an additional culture in fresh medium for 24h, 48h, 3 days, 7 days, and 14 days. Peripheral blood mononuclear cells (PBMC) were obtained from different sources. PBMC (SBK) was purchased from a biotechnology company (Saibaikang, China). PBMC (donors) was isolated from peripheral blood of four healthy donors. Informed consent was provided all the subjects. PBMC cells were cultured in RPMI-1640 (HyClone, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA), maintaining in an incubator with a humidified atmosphere of 5 % CO2. The physiologically relevant concentrations of potassium dichromate used here were selected according to previous studies in LCLs and a higher than 70% cell viability (Lou et al. 2013). Human LCLs were also exposed to Actinomycin D (AMD, 0.2μg/ml, an RNA Pol I inhibitor that disrupts nucleolar integrity), Cadmium (CdCl2, 5–20μM), 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB, 33 mg/ml, an RNA Polymerase II inhibitor, RNA Polymerase III Inhibitor 1 (30μM), 5-Aza-cytidine (10μM, a DNA methylation inhibitor), 5-Aza-2’-deoxycytidine (10μM, a DNA methylation inhibitor), and Cisplatin (20μM, a DNA damaging agent).
DNA isolation
A minimum of three biological replicates for each group were used for DNA isolation. Genomic DNA was isolated using QIAamp DNA Blood Mini Kit (Qiagen) and Wizard® Genomic DNA Purification Kit (Promega) following the manufacturer’s protocol. Low molecular weight (LMW) DNA was extracted with ZR BAC DNA Miniprep Kit (Zymo BAC) and Zyppy™ Plasmid Miniprep Kit (Zymo plasmid) following the manufacturer’s protocol. In addition, a Hirt solution was also used for LMW DNA isolation as previously described by Ziegler et al (Denham et al. 2014). Briefly, after rinsing cells with 125 μl of cold PBS, the cells were lysed by adding 125 μl of Hirt solution (1.2% SDS, 20Mm EDTA) without pipetting, incubated at room temperature for 10 min, at which point 250 μl of 2M NaCl was added followed by incubation overnight at 4°C. After centrifugation at 14000g for 40 min at 4°C, the supernatant was mixed with phenol:chloroform:isoamyl alcohol (25:24:1) by vortexing. The aqueous phase was transferred to a new tube after centrifugation at room temperature for 5 min at 16,000g, and 1μl of glycogen (20 μg/μl), 0.5 × volume of 7.5 M NH4OAc, and 2.5 × volume of 100% ethanol was added to the aqueous phase, placed the tube at –80°C overnight. DNA pellets were obtained by centrifuging at 4°C for 30 minutes at 16,000g, followed by washing DNA with 70% ethanol. DNA pellets were dried at 56°C for 1 min, re-suspended in AE buffer and preserved at −20°C. Total RNA was extracted using Trizol reagent (Invitrogen, USA) from cells before or after treatment with chromium. Purity and concentration of total RNA preparations was measured with a NanoDrop 2000 (Thermo Scientific, USA). Complementary DNA (cDNA) was synthesized from 500 ng of total RNA with First Strand Kit (Qiagen, Germany).
rDNA CN detected with Droplet digital PCR (ddPCR) and real time PCR (qPCR).
Droplet digital PCR (ddPCR) was performed according to the manufacture’s recommendation; Also see Miotke et al (Miotke et al. 2014). Briefly, digestion of gDNA with restriction enzyme Hind III overnight at 37°C was conducted as the initial step. The 20 μl of PCR reaction consisted of 2×EvaGreen ddPCR Supermix (BioRad), primers at the final concentration of 125 nM, and 1 ng of digested genomic DNA (gDNA). Approximately 20,000 droplets for each reaction mixture were generated with a droplet generator (BioRad QX200). The reaction was cycled with the following conditions: 5 min at 95°C (1 cycle); 30s at 95°C and 1 min at 60°C (40 cycles); 5 min at 4°C and 5 min at 90°C (1 cycle), 4°C hold. QX200 droplet-reader (BioRad) was used to read cycled droplets. The concentration of each target was calculated using QuantaSoft software, and the CN of each rDNA component was normalized by the single copy genes TP53 and CCND1. Real time quantitative PCR was performed to detect rDNA CN with KAPA SYBR FAST Universal PCR Master Mix (Kapa Biosystems) using 7900HT Fast Real-time PCR system (Applied Biosystems). Twenty nanograms of gDNA and 125 nM of each primer were used in PCR reactions (40 cycles of 10s at 95°C and 30 s at 60°C). Dissociation curves were used to assess unspecific amplification, and the data was normalized by delta CT values from single copy genes (TP53 and CCND1).
Immunofluorescence and confocal microscopy
After treatment, cells were fixed in methanol (5 minutes, RT), incubated in Image-iT FX signal enhancer (30 minutes, RT), and blocked in 3% BSA (1h, RT). Primary antibody against nucleolin (1/1000, Abcam) were diluted in 3% BSA and incubated overnight at 4°C. After washing in TBST, secondary antibody (green) Alexa Fluor® 488 goat anti-rabbit IgG (H+L) (Abcam) was used at 1/1000 dilution for 1h. At the same time, Alexa Fluor® 594 WGA (Abcam) was used to label plasma membrane (red). Images of cells were captured through a confocal microscope (Zeiss LSM) using a 63× lens, and the images were processed with Fiji software.
Data analysis in cell culture experiments
All representative CN data were from at least three independent experiments for each point. One-way ANOVA followed by LSD post hoc test (equal variances) or Dunnett’s T3 post hoc test (unequal variances) was used to analyze the differences among treatments. Student’s t-test was used to determine the significance of differences in rDNA CN between control and exposed groups. Statistical analyses of the cell culture experiments were performed with SPSS 11.0.
Drosophila exposure
Drosophila (strain 29656, DSPR panel) was kept at 23°C in vials filled with molasses medium (850 mL of water, 5 g of agar, 27.5 g of Torula yeast, 52 g of corn meal, 55 g of dextrose, and 40 mL of unsulfured molasses cooked at 95°C for 10 minutes). In order to generate a genetically homogeneous population in this inbred strain, individual crosses of single male and female were carried out for 5 generations immediately prior to the exposure experiment. Stability of the rDNA in the reference strain was checked periodically by qPCR and ddPCR. To evaluate the impact of Cr (VI) on rDNA stability, 2-day-old flies (5 males and 10 females) were transferred to new vials with food containing 0.25 mM of Cr (K2Cr2O7). After 3 days laying eggs, adult flies were discarded. Newly emerged male flies (F1) were collected daily to control for age and aged for 2 days in vials containing the same Cr-containing of food in which they were raised. At the end of day 2, flies were flash frozen in liquid nitrogen and stored at −80°C. This process was repeated for two additional generations to assess the effect of Cr (VI) on the F2 and F3 progeny. After 3 generations (F3) in food containing Cr (VI), flies were transferred to regular medium (without chromium) and raised for 2 additional generations (F4 and F5). The F4 and F5 progeny raised in regular food were collected and handled according to the method described above.
Drosophila DNA isolation and rDNA CN
Sets of 10 flies were carefully ground with small pestles in 1.5 mL tubes immersed in dry ice. After obtaining a fine powder, tubes were transferred to regular ice for 5 minutes and DNA isolated with QIAamp DNA Mini Kit (Qiagen). The DNA samples eluted in 50 μl of 50 ug/mL of RNAse-A were incubated at room temperature for 20 minutes to eliminate RNA, and DNA concentration was estimated with Nanodrop. PCR reactions were carried out with KAPA Sybr Fast Universal qPCR kit (KAPA Biosystems), 20 ng of gDNA, and 125 nM of primers to a final volume of 20 uL per reaction. Samples were submitted to Applied Biosystems 7900HT Fast Real-Time PCR System for 40 cycles (95°C for 10 seconds, and 60°C for 45 seconds). Relative CN of the 5S and 5.8S rDNA were estimated relative to the single-copy reference gene elF5.
Human populations
Ribosomal DNA estimates were obtained from short-read DNA sequencing (DNAseq) data. We estimated rDNA CN in the blood of 1,092 individuals from The Cancer Genome Atlas (TCGA) panel using methods previously described (Gibbons et al. 2015). Additionally, human peripheral blood samples were collected from 92 workers occupationally exposed to hexavalent chromium and 93 controls. The workers are electroplating workers; samples were collected at the end of the week. The characteristics of electroplating workers and controls are shown in Table 1. No significant difference of age, gender, smoking habits, and drinking consumption was found between these two groups. Our research was performed according to a protocol approved by the Ethics Committee of Zhejiang Academy of Medical Sciences (ZAMS). Written informed consents were obtained from subjects participating in this study. DNA samples were isolated from whole blood using NucleoSpin columns (Macherey-Nagel, Germany). rDNA CN was detected with ddPCR/qPCR as described above. Chromium concentrations in blood samples were measured with inductively coupled plasma-mass spectrometry (ICP-MS, PerkinElmer, USA) using aliquots of blood (0.2 ml) diluted with 5% v/v Tetramethylammonium hydroxide (TMAH) (Sigma–Aldrich, St. Louis, USA) solution. ICP-MS analyses were conducted after incubation at room temperature for 10 min and determined with isotopes of 52Cr. Association between variables was evaluated with Spearman rank correlation. Differences in continuous variables were evaluated using Student’s t-test and nonparametric two-sample Wilcoxon test.
Table1.
Characteristic of electroplating workers and controls. Blood concentration of Chromium (BCr, ug/L)
| Characteristic | Control | Exposed | Z/X2 | P |
|---|---|---|---|---|
|
| ||||
| (n=93) | (n=92) | |||
| Age | 43.16 ± 7.23 | 42.26 ± 9.09 | −0.839 | 0.401 |
| Gender | 0.959 | 0.327 | ||
| Male | 76 (81.7%) | 80 (87.0%) | ||
| Female | 17 (18.3%) | 12 (13.0%) | ||
| Smoking | −2.279 | 0.023 | ||
| No | 45 (48.4%) | 25 (27.2%) | ||
| Occasionally | 6 (6.5%) | 15 (16.3%) | ||
| Often | 42 (45.1%) | 52 (56.5%) | ||
| Drinking | −0.361 | 0.718 | ||
| No | 39 (41.9%) | 43 (46.7%) | ||
| Occasionally | 42 (45.2%) | 26 (28.3%) | ||
| Often | 12 (12.9%) | 23 (25.0%) | ||
| Exposure (Years) | - | 8.14 ± 5.88 | ||
| BCr, mean ± SD (Range) | 4.41 ± 3.63 (0.01–22.08) | 9.58 ± 15.44 (0.04–58.60) | −1.882 | 0.06 |
Results
Cr (VI) exposure induces a concentration-dependent amplification of the rDNA
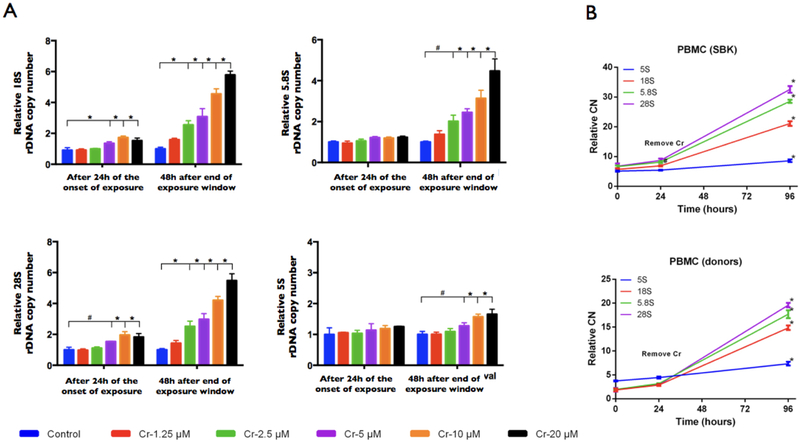
We first addressed whether a 24h exposure to physiologically relevant concentrations of Cr (VI) (K2Cr2O7) can induce rDNA CN instability (Fig. 1). To address the issue, we treated (t=0) human lymphoblastoid cell lines (LCL) with a series of Cr (VI) (potassium dichromate) concentrations for 24h (the interval from the start time at t=0 to t=1 at 24h is the exposure window, EW) (Fig. 1). Cr (VI) exposure did not induce significant CN changes in neither the 5S nor the 45S rDNA arrays during the EW interval. While CN was stable over the 24h interval of exposure, continued incubation of the cells in fresh medium revealed a delayed response, with all 45S rDNA components sharply amplified after an additional 48h culture in fresh medium (Fig. 2a; P < 0.05, Student’s t-test). The rDNA amplification occurred at all treatment concentrations and followed a simple dose-response relationship (Fig. 2a). Also, we observed modest but significant amplification of the unlinked 5S rDNA array at > 2.5 μM Cr (VI) (P < 0.05) (Fig. 2a). Finally, we addressed whether primary mononuclear blood cells isolated from normal individuals would be susceptible to Cr (VI)-induced rDNA-amplification. Accordingly, treating freshly isolated PMBCs with Cr (VI) induced significantly amplification of the rDNA (Fig. 2b) in a manner that mimicked the initial observations with immortalized LCLs. Thus, physiologically relevant concentrations of Cr (VI) induced significant rDNA array stability in human cells.
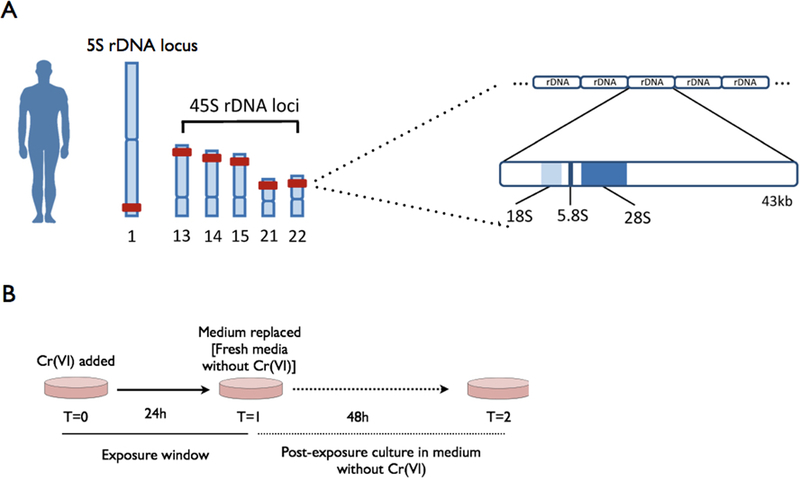
Figure 1.
Ribosomal DNA (rDNA) structure and environmental exposure. (a) In the human genome, the 45S rDNA unit (encoding the 18S, 5.8S and 28S rRNAs) is tandemly arrayed in nucleolar organizer regions (NORs) residing on five acrocentric chromosomes (13, 14, 15, 21 and 22). While the tandemly repeated cluster encoding the 5S rRNAs is located on chromosome 1. (b) Paradigm for hexavalent chromium [Cr (VI)] exposure.
Figure 2.
Hexavalent chromium [Cr (VI)] induces rDNA-amplification in human cells. (a) Dose dependent amplification of the rDNA array in genomic DNA from human LCLs exposed to hexavalent chromium [Cr (VI)]. Dose response relationships from exposure with the following concentrations of Cr (VI): 1.25 μM, 2.5 μM, 5 μM, 10 μM and 20 μM. Shown are rDNA CN estimates prior to the onset of exposure, immediately after the 24h exposure window (t = 1), and after continued culture for 48h after the end of the Cr (VI) exposure window, EW [t = 2, 48h after the end of the EW)]. Copy number of the 5S and 45S (18S, 5.8S, 28S) rDNA components were normalized with single copy genes TP53 and CCND1 (qPCR and ddPCR). Bars denote standard deviation; fold changes are shown relative to unexposed control (* P < 0.01; # P < 0.05). (b) Exposure to hexavalent chromium [Cr (VI), 5 μM] induces rDNA amplification in primary human cells. Peripheral blood mononuclear cells (PBMC) were isolated from commercially obtained primary mononuclear cells [PBMC (SBK)] or anticoagulated blood from donors [PBMC (donors)]. Cells were treated with 5 μM of K2Cr2O7 for 24h, transferred to fresh medium, and incubated for additional 3 days without chromium. rDNA CN estimates as in (a) above (mean of 45S components is shown).
Cr (VI) exposure induces nucleolar fragmentation
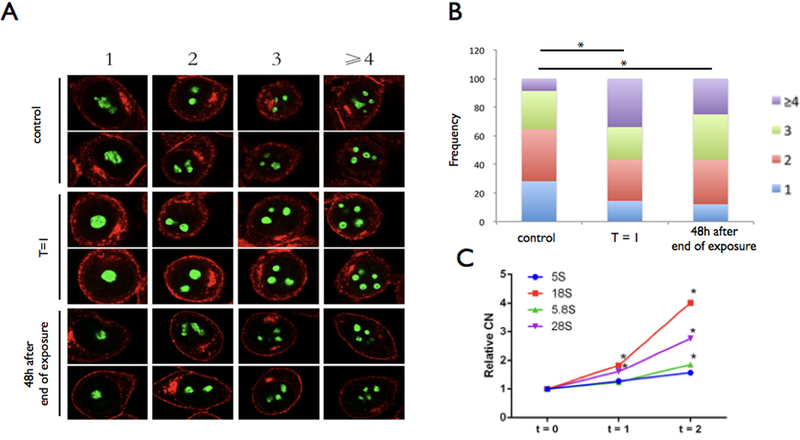
Nucleoli fragmentation has been observed before with classical inducers of nucleolar stress (e.g, Actynomycin D, AMD). Thus, we monitored nucleolar changes in human HeLa cells upon Cr (VI) exposure. The cells showed significant nucleolar fragmentation that was detectable during the 24h exposure window and rDNA amplification in the subsequent 48h after passage to fresh medium (Fig. 3, P < 0.01, Student’s t-test). The percentage of cells with one or two nucleoli decreased during or after Cr (VI) treatment, while the percentage of cells with three or more nucleoli increased significantly (Fig. 3; P < 0.01, Chi-square test). Thus, Hela cells displayed nucleolar fragmentation and qualitatively similar dynamics of rDNA amplification after continued culture as we had observed for LCLs and PBMCs.
Figure 3.
Hexavalent chromium [Cr (VI)] induces nucleolar fragmentation. (a, b) Treatment with 5 μM in Hela cells; t=1 and t=2 as in Figure 2. t=0 is the control. Images were captured through a confocal microscope (Zeiss LSM) using a 63× lens; images were processed with the Fiji software. Nucleolus (green) stained with antibody against nucleolin were counted across 3 independent experiments (>50 cells in each replicate). Plasma membrane stained in red. Number of nucleoli counted for >75 cells in each treatment (* P < 0.01, Fisher’s exact test). (c) rDNA CN estimates as in figure 2 (mean of 45S components is shown).
Blocking DNA replication attenuates Cr (VI) induced rDNA amplification
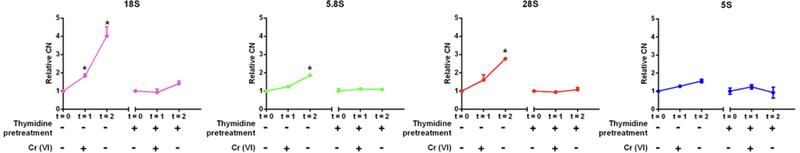
Because of the delayed rDNA CN amplification upon Cr (VI) exposure, we hypothesized that amplification was dependent on cell cycle progression. In order to investigate this hypothesis, we synchronized HeLa cells with thymidine, an agent that blocks DNA synthesis and cell cycle progression. Cells with or without thymidine pre-treatment were exposed to Cr (VI) for 24h. As in prior protocols, we removed the Cr (VI) containing media at the end of the 24h exposure window and continued with another 48h incubation of cells in fresh medium. The results further confirmed that Cr (VI) exposure induced rDNA CN in HeLa cells without thymidine treatment. Copy number of the 18S and 28S rDNA 48h after removal of Cr (VI) was amplified about two times above the baseline at the end of the exposure window (P < 0.01). However, upon thymidine treatment, Cr (VI) exposure failed to induce a significant increase of rDNA CN in any group (Fig. 4).
Figure 4.
Blocking DNA synthesis prevents Cr (VI)-induced rDNA amplification. Double thymidine treatment attenuated Cr (VI) induced rDNA amplification. rDNA CN was detected in genomic DNA samples from HeLa cells (with or without a double thymidine pre-treatment) before (t=0) and after exposure to 5μM Cr (VI) for 24h (t=1), and from cells after continued culture for 48h (t=3). rDNA CN were normalized by the control at each time point. Symbols denote a significant difference between control and treatment (* P < 0.01; # P < 0.05).
rDNA CN is robust to treatment with a variety of chemical inhibitors
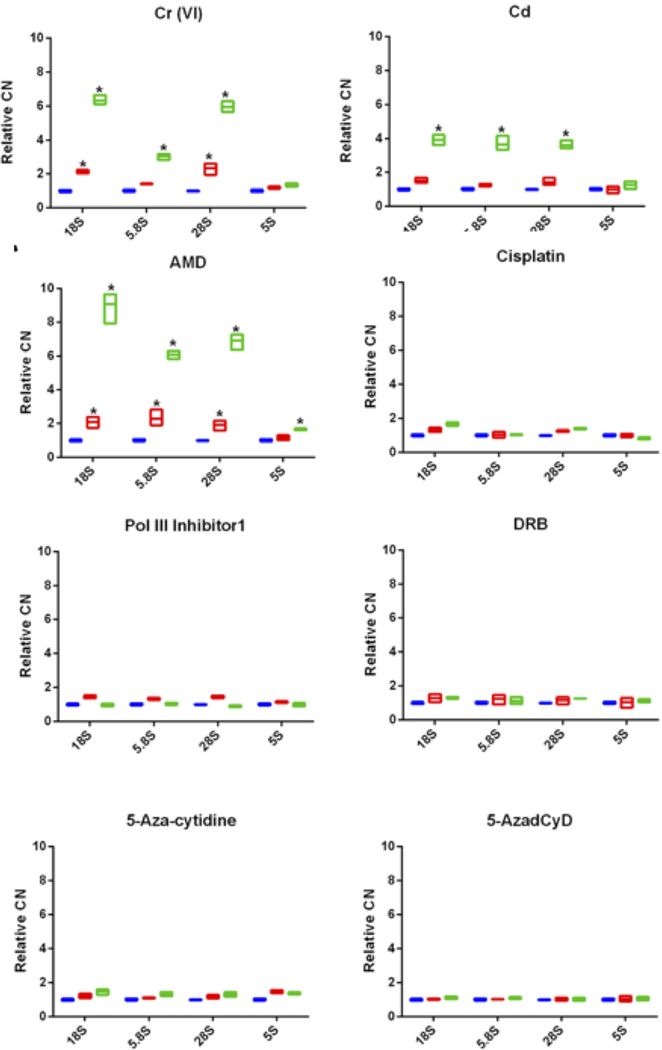
Chemotherapeutic drugs are known to impact the ribosome biogenesis and nucleolar integrity, and the effects on the integrity of nucleus rely on the blockage of ribosomal biogenesis at early or late stage by inhibiting rRNA expression (Burger et al. 2010). Here we investigated the effects of five chemicals inhibitors on ribosomal DNA CN. As expected, LCL treatment with 0.08 μM of Actinomycin D (AMD), which is known to inhibit rRNA transcription in HeLa cells after 4μM exposure for 3h (Cong et al. 2014), significantly elevated rDNA CN. The rDNA amplification induced by AMD was higher than that induced by Cr (VI). On the other hand, neither Pol II or Pol III inhibitors nor a widely used DNA damaging agent (cisplatin) induced changes in rDNA CN (Fig. 5). These data indicate that inhibition of RNA polymerase I might be associated with rDNA amplification, whereas inhibition of RNA polymerase II or III does not impact rDNA CN. Finally, treatment with two DNA methyltransferase inhibitors that result in genome-wide demethylation (5-Aza-cytidine and 5-Aza-2’-deoxycytidine) did not induce changes in rDNA CN (Fig. 5).
Figure 5.
rDNA CN is stable to treatment with a variety of chemical inhibitors. Human LCLs were exposed to Cr (VI) (5μM), Cadmium (CdCl2, 10μM), Actinomycin D (AMD, 0.2μg/ml), Cisplatin (20μM), RNA Polymerase III Inhibitor (30μM), 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB, 33 mg/ml), 5-Aza-cytidine (10μM), and 5-Aza-2’-deoxycytidine (10μM). rDNA CN immediately prior to the onset of exposure (t=0, denoted in blue). In all cases cells were exposed for 24h (t=1 is the end of the exposure window, denoted in red). Cells were further incubated in fresh medium for an additional 48h after the end of the exposure window (t=2, denoted in green). Fold changes are shown relative to unexposed control. Asterisks denote a significant difference between control and treatment (* P < 0.01).
Cr (VI) induced rDNA amplification did not induce LMW rDNA fragments
In yeast, the rDNA arrays are known to produce low molecular weight circular molecules as well as fragments that are presumably produced during replication and transcription (Moller et al. 2015; Mansisidor et al. 2018). However, the prevalence of low molecular weight rDNA has remained uncertain in humans. Hirt extraction is the method of choice to identify low molecular weight (LMW) DNA emerging from the nuclear genome. Hence, to access LMW rDNA we used Hirt extraction as well as two kits [ZR BAC DNA Miniprep Kit (Zymo BAC), Zyppy™ Plasmid Miniprep Kit (Zymo plasmid)] for the isolation of plasmid DNA. These were contrasted with two procedures to isolate genomic DNA [QIAamp DNA Blood Mini Kit (Qiagen) and Wizard® Genomic DNA Purification Kit (Promega)]. Noteworthy, quantification of rDNA CN by qPCR and ddPCR yielded relative estimates that were highly correlated (rho= 0.93 for 5S, rho= 0.97 for 18S, rho= 0.92 for 5.8S, rho= 0.92 for 28S, P < 0.01).
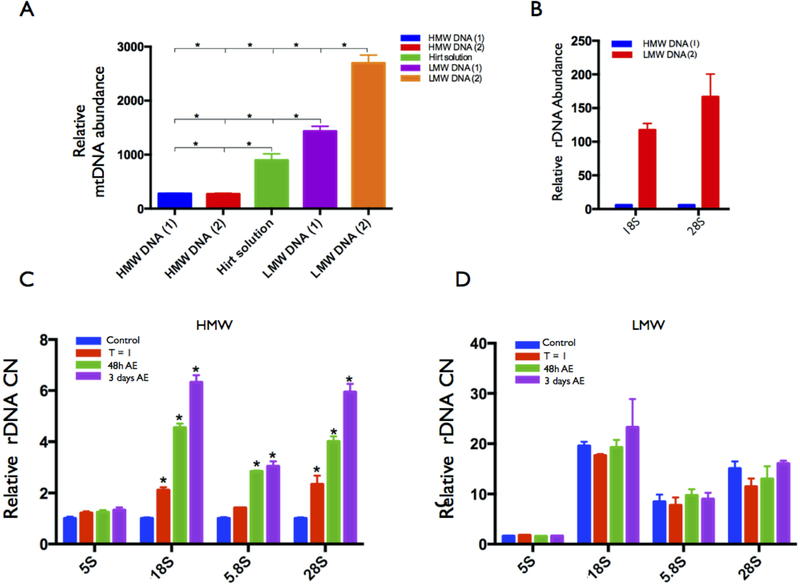
We used estimates of mtDNA CN in each extraction method as an indicator of the efficiency to isolate high and low molecular weight DNA. Our results showed similarly low mtDNA CN estimates in DNA extracted with Qiagen and Promega kits (Fig 6a), which indicate enrichments in high-molecular weight DNA. On the other hand, mtDNA CN in LMW DNA was substantially higher (P < 0.01), with over 3000-fold enrichment in mtDNA in LMW pool relative to HMW DNA (Fig. 6a). In particular, LMW isolated with Zymo-BAC kit showed the highest mtDNA CN estimate. Hence, we assessed the differential representation of rDNA copies in HMW vs LMW pools using Qiagen kit and Zymo-BAC to isolate genomic HMW DNA and LMW DNA, respectively. As expected, both qPCR and ddPCR (Fig. 6b) showed that, relative to single copy nuclear genes, the number of rDNA molecules was significantly higher in LMW than in HMW DNA (P < 0.01) (ratio of LMW rDNA/LMW single copy genes >> HMW rDNA/HMW single copy genes). This is partially because of a small denominator due to single copy genes being negligeable small in the LMW pool. However, we note that rDNA copies in LMW DNA pool were also significantly smaller than in the HMW DNA pool (P < 0.01).
Figure 6.
Cr (VI) exposure does not increase the abundance of low molecular weight rDNA fragments. (a) DNA isolation and estimates of LMW DNA in human LCLs. Mitochondrial DNA (mtDNA) abundance was detected (qPCR and ddPCR) from DNA isolated with Qiagen genomic DNA kit (HMW1), Promega genomic DNA kit (HMW2), Zymo plasmid kit (LMW1), Hirt solution (LMW2), and Zymo Bac kit (LMW3); Zymo plasmid kit, Hirt solution, and Zymo Bac kit significantly enriched for LMW mtDNA. Estimates of mtDNA abundance with a primer targeting the mitochondrial COX1 gene and normalized with the single copy genes TP53 and CCND1. (b) rDNA abundance detected in HMW and LMW DNA. Data for 18S and 28S components was normalized with the single copy genes TP53 and CCND1. (c) Comparing the rDNA CN induced by Cr (VI) between genomic DNA (gDNA) and low molecular weight DNA (LMW DNA). The rDNA CN was detected with qPCR in genomic DNA (c) and low molecular weight DNA (d) from Human LCL cells after being exposed to 10 μM Cr (VI) for 24h [Cr(VI)-24h] and from cells with an additional incubation of 24h [Cr(VI)-24R] or 48h [Cr(VI)-48R] after removing Cr (VI). Data was normalized by the single copy gene TP53. Asterisks represent a significant difference between groups (P < 0.01).
Next, we addressed the potential for Cr (VI) to differentially affect the pools of high and low molecular weight rDNA. To address the issue, we exposed LCLs to 5 μM of Cr (VI) for 24h followed by additional 24h and 48h incubation in fresh medium after removal of Cr (VI). As previously observed, Cr (VI) induced rDNA CN amplification in genomic DNA (Figs. 2–5). However, analysis of LMW DNA pool indicated that rDNA abundance in LMW DNA remained stable during the same period (Fig. 6d). Thus, we conclude that while the abundance of rDNA relative to single copy DNA was much higher in LMW DNA than in HMW genomic DNA preps, Cr (VI) exposure did not increase the prevalence of low-molecular weight rDNA fragments.
Cr-induced rDNA-amplification is transient and followed by rapid contraction.
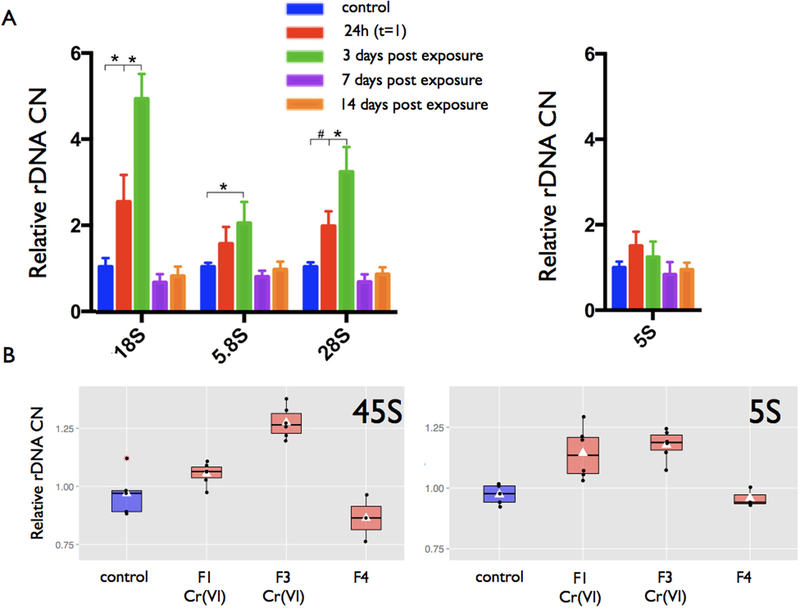
To investigate the long-term effects of Cr (VI) on rDNA CN, we monitored the dynamics of Cr (VI) induced rDNA amplification over a two-week interval. Based on results from the concentration-response curves and empirical estimates of Cr concentration in human blood, we chose 5 μM of Cr (VI) as the concentration of choice. Following the same exposure paradigm, human LCLs were treated with Cr (VI) for 24h. Cr (VI) was removed at 24h after the onset of exposure and cells were maintained with fresh medium for an additional 2 weeks. rDNA CN was detected at five time points: prior to exposure (t=0), immediately after the exposure window (t=1), 72h after the end of the exposure window (t=3), 7 days after the end of the exposure window (t=4), and 2 weeks after the end of the exposure window (t=5). We observed a peak in rDNA CN three days after the end of the exposure window (Fig. 7). Surprisingly, however, rDNA CN in exposed cells returned to the level of the control group within one week after removal of Cr (VI) (Fig. 7). To further investigate this, we assayed adult Drosophila exposed to Cr (VI). Exposure occurred throughout development as unexposed adults layed eggs into Chromium containing food where flies developed from egg to larvae and into adults. We observed a significant increase in rDNA CN in flies exposed to Cr (VI) throughout their development (P < 0.01, Student t-test). The magnitude of the amplification became more pronounced upon multiple consecutive generations of exposure (Fig. 7). However, and in agreement with human cell experiments, relief from Cr (VI) exposure by transferring exposed flies to lay eggs in regular food caused rDNA CN values to return to their original estimates in a single generation.
Figure 7.
Cr (VI) induced rDNA amplification is transient. Temporal dynamics of rDNA amplification and subsequent contraction following an event of Cr (VI) exposure in LCLs. (a) rDNA CN estimates from genomic DNA before and after exposure to 5 μM Cr (VI) for 24h (24h) and from cells with an additional post exposure incubation period of three days (3d), seven days (7d), and two weeks (14d) after the removal of Cr (VI). Relative CNs are normalized by the control at each time point, and the bars in each represent the standard deviation. Symbols denote a significant difference between control and treatment (* P < 0.01; # P < 0.05). (b) Cr (VI) induced rDNA CN amplification in Drosophila. Parental flies layed eggs in food containing 0.25 mM of Chromium (K2Cr2O7). After 3 days laying eggs, adult flies were discarded. Newly emerged flies (F1) were collected daily and genotype or replicated for continued culture in Cr (VI) containing food. A set of F3 adults were returned to regular food for egg laying. F4 flies developed in regular food without Cr (VI).
Extensive naturally occurring rDNA CN is partially modulated by exposure history.
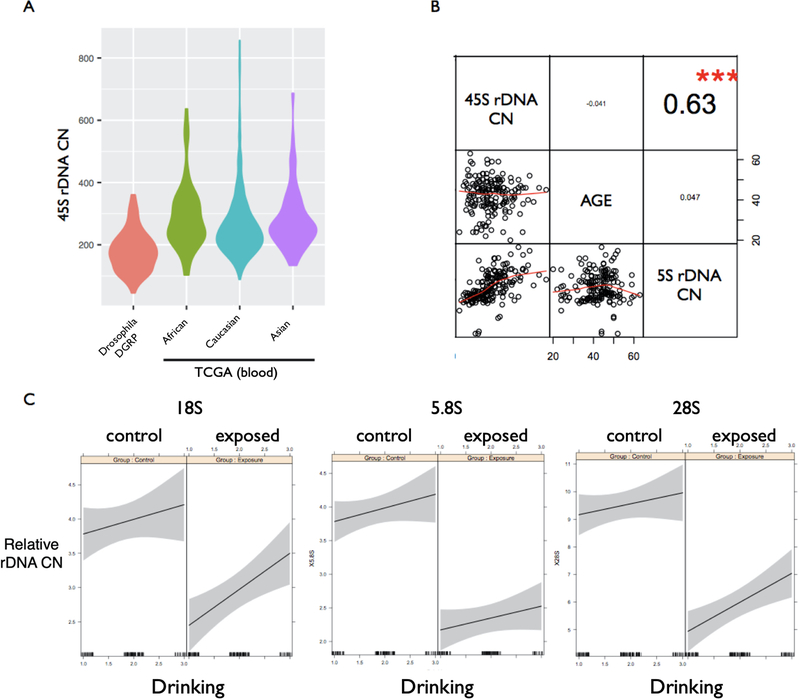
Here we further revisited the extent of rDNA CN in natural human populations using short DNA sequencing reads (Gibbons et al. 2014) to genotype blood samples of 1,092 individuals from the TCGA panel. The data confirmed extensive rDNA CN diversity in humans (Fig. 8; Mean = 538 copies per diploid genome; range = 131–1,070 copies); substantial rDNA CN variation is also detected across 205 inbred Drosophila genotypes of the Drosophila Genetic Reference Panel (DGRP) (Fig. 8). Noteworthy, we found no association between rDNA CN and age in humans. Next, we addressed the hypothesis that rDNA CN is partially linked to Cr (VI) exposure history in a population with chronic occupational exposure to metals. We evaluated rDNA CN in 92 workers occupationally exposed to Cr (VI) and other metals and in a control group of 93 individuals not occupationally exposed to Cr (VI) and matched for age, gender, smoking habits, and drinking consumption. As expected, the exposed population had higher levels of Cr (VI) in blood (9.58μg/L in exposed workers vs 4.41μg/L in non-exposed controls, P < 0.01, Student’s t-test). Surprisingly, 45S rDNA CN was significantly lower in the exposed population (Fig. 8) for all components tested (P < 0.01, in all three components). Similarly, the 5S rDNA was also significantly lower in the exposed population (5S: 3.46±0.79 in exposed workers vs 4.83±0.84 in the non-exposed control, P < 0.01). Finally, neither age nor smoking habits were associated with rDNA CN (Fig. 8), whereas a significant positive association between rDNA CN and drinking habits is manifested in both exposed and non-exposed populations (Fig. 8). Collectively, the data indicate that environmental exposure history can partially explain the extensive diversity of rDNA CN in natural populations.
Figure 8.
Extensive ribosomal DNA CN variation in human populations is associated with exposure history but not age. (a) Violin plots show 45S rDNA CN variation in fruit flies (205 genotypes of the DGRP panel) and humans (1,092 individuals from the TCGA panel with African, Caucasian, and Asian ancestry; DNA from whole blood). (b) rDNA CN variation in a population of Chinese workers occupationally exposed to Cr (VI) and controls demographically matched and sampled in the same city. Copy number of the 5S and 45S are correlated with each other, whereas neither 5S CN nor 45S CN are correlated with individual age. (c) rDNA CN is significantly reduced in individuals occupationally exposed to Cr (VI) (right panel) relative to controls (left panel) in each rDNA component assessed (P < 0.001), while positively associated with drinking habits (X-axis; 0 = no drinking, 1 = occasional drinking, 3 = regular drinking) in both control (left panel) and exposed (right panel) populations (grey areas represent 95% confidence intervals for the linear coefficients).
Discussion
Exposure to environmental toxicants is ubiquitous through the life-course. For instance, considerable levels of chromium, lead, and cadmium is in the drinking water and food sources of millions of individuals worldwide (IARC 1990; Mancuso 1997a; Mancuso 1997b; Smith and Steinmaus 2009). Occupational exposure in pigment factories or welding environments cause substantially higher daily exposures that result in individuals displaying blood Cr (VI) concentration that can be >5–10x higher than those found in the general population (IARC 1990; Mancuso 1997a; Mancuso 1997b; Smith and Steinmaus 2009). Exposure to Cr (VI) at physiologically relevant doses for 24h induced a delayed concentration-dependent rDNA amplification response. Once triggered, the amplification process was sustained for at least three days after removal of Cr (VI) with progressive increases in CN during the interval. Surprisingly, wild-type rDNA CN was eventually restored upon continued culturing: the rDNA CN was amplified, peaked, and returned to wild-type levels within 7 days of exposure. Restoration of the original rDNA CN upon removal of the exposure might be partially due to genetic determinants of rDNA CN.
The rDNA array gives origin to the nucleolus, a crucial organelle sensing cellular stress and coordinating metabolism, energy status, and nuclear architecture (Pederson 1998; Boulon et al. 2010; O’Sullivan et al. 2013; Yu and Lemos 2018). Copy number of the rDNA has an outsize importance to nucleolus function and is intricately linked to overall cell physiology, proliferation, genome integrity, and genome-wide gene expression (Ide et al. 2010; Gibbons et al. 2014; Wang and Lemos 2017). Links between rDNA CN and global gene expression control have been suggested to partially reflect a regulatory architecture that functions to maintain a balanced stoichiometry of protein and RNA components of the ribosome (Birchler et al. 2005; Birchler and Veitia 2012; Gibbons et al. 2015; Birchler et al. 2016; Yu and Lemos 2018).
Environmental modulation of rDNA CN further contributes to naturally occurring rDNA diversity. Millions of individuals worldwide are regularly exposed to chromium and other metals through drinking water, diet, or via occupational exposure. Cr (VI) undergoes intracellular non-enzymatic reduction to Cr (III), a very toxic species that causes genetic damage (Shi et al. 1999; Liu and Shi 2001; Costa and Klein 2006). Upon exposure, multiple cellular pathways contribute to repair of chromium damaged DNA: mismatch repair (MMR), transcription-coupled nucleotide excision repair (TCR), DNA double-strand breaks (DSB) repair, among others (Hartwig et al. 2002; Hartwig and Schwerdtle 2002; Wise et al. 2008). One mechanism of damage is through the formation of Cr-DNA adducts that are initiated by Cr (III) and that preferentially occur at GG di-nucleotides (Arakawa et al. 2012). Such Cr-DNA interactions are likely to impact DNA repair processes that are specific to the nucleolus (van Sluis and McStay 2015; Warmerdam et al. 2016).
Low molecular weight (LMW) DNA in cell-free blood samples has long been linked to pathological conditions including carcinogenesis (Belokhvostov and Zelenkova 1978; Stroun et al. 1987), prenatal diseases (Lo et al. 1999), stroke (Rainer and Lam 2006), and xeroderma pigmentosum (Hurt et al. 1983). Elevated levels of LMW DNA are associated with genomic instability, aging, and cancer (Sinclair and Guarente 1997; Cohen and Lavi 2009) and has suggested to be toxic to cells (Sinclair and Guarente 1997). The formation of LMW DNA from the nuclear genome may occur through excision of chromosomal sequences (Cohen and Lavi 2009), rejoining of ends of fragmented DNA (van Loon et al. 1994), and homologous recombination (Stanfield and Helinski 1986; Cohen et al. 2008; Moller et al. 2015). It partially originates from the genome as extrachromosomal circular DNA (eccDNA). Here we investigated the rDNA in LMW DNA isolated with Hirt assays and commercial kits. We used mtDNA abundance as a control to evaluate methods to isolate LMW DNA. We observed that the relative abundance of the rDNA compared to single copy sequences was much higher in LMW DNA preparations than that in HMW genomic DNA. The increased relative abundance of rDNA in LMW DNA as compared to genomic DNA is expected because repetitive sequences are shed from internal chromosomal locations during replication in normal cells. However, here we observed that Cr (VI) did not affect the abundance of rDNA CN in LMW DNA fraction which remained stable in our study. The data argues against a model in which increased amount of LMW DNA may be either responsible for or a byproduct of Cr (VI) induced rDNA amplification.
Intriguingly, populations experiencing long-term exposure to Cr (VI) in the workplace displayed lower rDNA CN. While the dose and temporal variation in Cr (VI) exposure in these individuals is unknown, most of them have been exposed to metal mixtures over several years. Noteworthy, both human cells and Drosophila manifested a slightly lower CN immediately after contraction relative to the value prior to exposure, although the loss was minimal and not statistically significant in our study. Conceivably, chronically exposed individuals have experienced multiple cycles of rDNA expansion and contraction, with rDNA loss emerging as the net result of exposure over a prolonged period. Intriguingly, rDNA instability and net rDNA loss is also observed in cancers (Stults et al. 2009; Wang and Lemos 2017; Xu et al. 2017). Finally, the data also revealed an unexpected but significant positive association between rDNA CN and alcohol drinking habits in both exposed and control populations. The observations point to dynamic expansions and contractions that can be reconciled with evidence that rDNA CN is not in linkage with single nucleotide polymorphisms at segments flanking the array (Gibbons et al. 2015). One interpretation we previously advanced is that rDNA CN within individuals is constrained by stoichiometric demands and developmentally established with genetic and environmental determinants (Gibbons et al. 2015; Wang and Lemos 2017). Collectively, the data suggest a complex population dynamic of environmentally responsive rDNA CN variation.
Complex quantitative phenotypes are the result of genetic and environmental factors that manifest themselves through epigenetic processes (Waddington 1942a; Waddington 1942b; Visscher et al. 2008; Slatkin 2009; Furrow et al. 2011; Gibson 2012; Boyle et al. 2017). Rapid and reversible heritable responses to the environment over short timescales typically emerge through dynamic epigenetic chemical modifications on the DNA or protein molecules. However, multicopy DNA arrays can also be exceptionally dynamic with mutation rates that have challenged allelic characterization even in parent-offspring trios (Jeffreys et al. 1985b; Jeffreys et al. 1985a; Jeffreys et al. 1988; Stults et al. 2008). Evidently, an exquisitely labile rDNA genotype could contribute to heritability components, but the locus ability to be environmentally modulated present both conceptual and analytical challenges. This is because rDNA CN behaves as a polygenic quantitative trait that is dynamically modulated on the rapid timescale expected for epigenetic modifications. Thus, clear-cut distinctions between genetic and environmental influences on organismal phenotypes are bound to be blurred in the context of environmentally responsive rDNA elements.
Acknowledgments
We thank several members of the Lemos laboratory for their comments and feedback during development of this study and four anonymous reviewers for their comments on an early draft of the manuscript. Work in the Lemos laboratory has been supported by NIEHS grants R01ES027981 and P30ES000002. This research was also partially supported by a Harvard Global Institute (HGI) award.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Arakawa H, Weng MW, Chen WC, Tang MS. 2012. Chromium (VI) induces both bulky DNA adducts and oxidative DNA damage at adenines and guanines in the p53 gene of human lung cells. Carcinogenesis 33: 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, Costa M. 2009. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics 1: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi D, Stohs SJ, Downs BW, Bagchi M, Preuss HG. 2002. Cytotoxicity and oxidative mechanisms of different forms of chromium. Toxicology 180: 5–22. [DOI] [PubMed] [Google Scholar]
- Belokhvostov AS, Zelenkova NK. 1978. Low-molecular weight nucleic acids in the blood of rats with Zajdela’s hepatoma. Cancer Lett 5: 351–356. [DOI] [PubMed] [Google Scholar]
- Bianchi V, Dal Toso R, Debetto P, Levis AG, Luciani S, Majone F, Tamino G. 1980. Mechanisms of chromium toxicity in mammalian cell cultures. Toxicology 17: 219–224. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Johnson AF, Veitia RA. 2016. Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci 245: 128–134. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. 2005. Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169: 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Holzel M et al. 2010. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 285: 12416–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Shi X. 2002. Intracellular signal transduction of cells in response to carcinogenic metals. Crit Rev Oncol Hematol 42: 105–121. [DOI] [PubMed] [Google Scholar]
- Cohen MD, Kargacin B, Klein CB, Costa M. 1993. Mechanisms of chromium carcinogenicity and toxicity. Crit Rev Toxicol 23: 255–281. [DOI] [PubMed] [Google Scholar]
- Cohen S, Houben A, Segal D. 2008. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J 53: 1027–1034. [DOI] [PubMed] [Google Scholar]
- Cohen Z, Lavi S. 2009. Replication independent formation of extrachromosomal circular DNA in mammalian cell-free system. PLoS One 4: e6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong R, Das S, Douet J, Wong J, Buschbeck M, Mongelard F, Bouvet P. 2014. macroH2A1 histone variant represses rDNA transcription. Nucleic Acids Res 42: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Klein CB. 2006. Toxicity and carcinogenicity of chromium compounds in humans. Crit Rev Toxicol 36: 155–163. [DOI] [PubMed] [Google Scholar]
- Denham J, Marques FZ, Charchar FJ. 2014. Leukocyte telomere length variation due to DNA extraction method. BMC Res Notes 7: 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrow RE, Christiansen FB, Feldman MW. 2011. Environment-sensitive epigenetics and the heritability of complex diseases. Genetics 189: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb HJ, Lees PS, Pinsky PF, Rooney BC. 2000. Lung cancer among workers in chromium chemical production. Am J Ind Med 38: 115–126. [DOI] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. 2015. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci U S A 112: 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Yu S, Lemos B. 2014. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat Commun 5: 4850. [DOI] [PubMed] [Google Scholar]
- Gibson G 2012. Rare and common variants: twenty arguments. Nat Rev Genet 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A, Asmuss M, Ehleben I, Herzer U, Kostelac D, Pelzer A, Schwerdtle T, Burkle A. 2002. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect 110Suppl 5: 797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A, Schwerdtle T. 2002. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett 127: 47–54. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Eicher EM, Yu MT, Atwood KC. 1974. The chromosomal location of ribosomal DNA in the mouse. Chromosoma 49: 155–160. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. 1972. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A 69: 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, Hu W, Rothman N, Klugman M, Weinstein SJ, Virtamo JR, Albanes D, Cawthon R, Lan Q. 2019. Variation in ribosomal DNA copy number is associated with lung cancer risk in a prospective cohort study. Carcinogenesis 40: 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt MM, Beaudet AL, Moses RE. 1983. Stable low molecular weight DNA in xeroderma pigmentosum cells. Proc Natl Acad Sci U S A 80: 6987–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. 1990. International Agency for Research on Cancer (1990) Chromium, nickel and welding. IARC [Google Scholar]
- Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 49, pp 49–256, World Health Organization, Lyon, France. [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T. 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Royle NJ, Wilson V, Wong Z. 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332: 278–281. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. 1985a. Hypervariable ‘minisatellite’ regions in human DNA. Nature 314: 67–73. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. 1985b. Individual-specific ‘fingerprints’ of human DNA. Nature 316: 76–79. [DOI] [PubMed] [Google Scholar]
- Langard S 1990. One hundred years of chromium and cancer: a review of epidemiological evidence and selected case reports. Am J Ind Med 17: 189–215. [DOI] [PubMed] [Google Scholar]
- Liu KJ, Shi X. 2001. In vivo reduction of chromium (VI) and its related free radical generation. Mol Cell Biochem 222: 41–47. [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. 1999. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 64: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB. 1980. Repeated genes in eukaryotes. Annu Rev Biochem 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Lou J, Wang Y, Yao C, Jin L, Wang X, Xiao Y, Wu N, Song P, Song Y, Tan Y et al. 2013. Role of DNA methylation in cell cycle arrest induced by Cr (VI) in two cell lines. PLoS One 8: e71031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lyckegaard EM, Clark AG. 1989. Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A 86: 1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso TF. 1997a. Chromium as an industrial carcinogen: Part I. Am J Ind Med 31: 129–139. [DOI] [PubMed] [Google Scholar]
- Mancuso TF. 1997b. Chromium as an industrial carcinogen: Part II. Chromium in human tissues. Am J Ind Med 31: 140–147. [DOI] [PubMed] [Google Scholar]
- Mansisidor A, Molinar T Jr., Srivastava P, Dartis DD, Pino Delgado A, Blitzblau HG, Klein H, Hochwagen A 2018. Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles. Mol Cell 72: 583–593 e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2014. Cellular Stress and Nucleolar Function. Cell Cycle 4: 1036–1038. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157. [DOI] [PubMed] [Google Scholar]
- Miotke L, Lau BT, Rumma RT, Ji HP. 2014. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal Chem 86: 2618–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller HD, Parsons L, Jorgensen TS, Botstein D, Regenberg B. 2015. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A 112: E3114–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JM, Pai DA, Cridge AG, Engelke DR, Ganley AR. 2013. The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear structure? Biomol Concepts 4: 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 1998. The plurifunctional nucleolus. Nucleic Acids Res 26: 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer TH, Lam NY. 2006. Circulating nucleic acids and critical illness. Ann N Y Acad Sci 1075: 271–277. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Peterson E, Quievryn G, Zhitkovich A. 2004. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J Biol Chem 279: 30419–30424. [DOI] [PubMed] [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F. 1993. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol 175: 7189–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. 2008. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol 21: 28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chiu A, Chen CT, Halliwell B, Castranova V, Vallyathan V. 1999. Reduction of chromium(VI) and its relationship to carcinogenesis. J Toxicol Environ Health B Crit Rev 2: 87–104. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Slatkin M 2009. Epigenetic inheritance and the missing heritability problem. Genetics 182: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Steinmaus CM. 2009. Health effects of arsenic and chromium in drinking water: recent human findings. Annu Rev Public Health 30: 107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage DE, Eickbush TH. 2007. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res 17: 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield SW, Helinski DR. 1986. Multiple mechanisms generate extrachromosomal circular DNA in Chinese hamster ovary cells. Nucleic Acids Res 14: 3527–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. 1987. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 23: 707–712. [DOI] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ. 2008. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. 2009. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res 69: 9096–9104. [DOI] [PubMed] [Google Scholar]
- Tully DB, Collins BJ, Overstreet JD, Smith CS, Dinse GE, Mumtaz MM, Chapin RE. 2000. Effects of arsenic, cadmium, chromium, and lead on gene expression regulated by a battery of 13 different promoters in recombinant HepG2 cells. Toxicol Appl Pharmacol 168: 79–90. [DOI] [PubMed] [Google Scholar]
- van Loon N, Miller D, Murnane JP. 1994. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res 22: 2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluis M, McStay B. 2015. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev 29: 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet 9: 255–266. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942a. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942b. The epigenotype. Endeavour 1–18. [Google Scholar]
- Wang M, Lemos B. 2017. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet 13: e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, van den Berg J, Medema RH. 2016. Breaks in the 45S rDNA Lead to Recombination-Mediated Loss of Repeats. Cell Rep 14: 2519–2527. [DOI] [PubMed] [Google Scholar]
- Wise SS, Holmes AL, Wise JP Sr. 2008. Hexavalent chromium-induced DNA damage and repair mechanisms. Rev Environ Health 23: 39–57. [DOI] [PubMed] [Google Scholar]
- Xu B, Li H, Perry JM, Singh VP, Unruh J, Yu Z, Zakari M, McDowell W, Li L, Gerton JL. 2017. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet 13: e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Lemos B. 2018. The long-range interaction map of ribosomal DNA arrays. PLoS Genet 14: e1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]