Abstract
Background
The role of the gastric microbiome in development or persistence of equine glandular gastric disease (EGGD) remains to be investigated.
Hypothesis/Objectives
The objective was to characterize the glandular mucosal and gastric fluid microbiomes of horses with and without EGGD. It was hypothesized that differences in the mucosal microbiome are associated with EGGD.
Animals
Twenty‐four horses were enrolled.
Methods
Gastroscopy was performed and EGGD scores recorded (score 0, n = 6; score 1, n = 8; score ≥2, n = 10). Gastric fluid and pinch biopsies of healthy glandular mucosa and EGGD lesions were collected via gastroscope. 16S rRNA amplicon sequencing of the gastric fluid and glandular mucosal biopsies was performed. Relationships between gastric fluid and mucosal microbial community composition were evaluated among EGGD score groups (EGGD 0‐BX, EGGD 1‐BX, EGGD ≥2‐BX) and among endoscopic appearances: controls from horses without EGGD and normal areas, hyperemic areas, and lesions from horses with EGGD.
Results
Microbial community structure of mucosal biopsies differed among EGGD score groups (Jaccard similarity index; P = .009). Principal coordinate analysis showed separate clusters for EGGD 0‐BX and EGGD ≥2‐BX.
Conclusions and Clinical Importance
A modest difference was detected in the community structure of the gastric glandular mucosal microbiome in association with EGGD score.
Keywords: bioinformatics, colic, equine gastric ulcer syndrome (EGUS), stomach
Abbreviations
- EGGD
equine glandular gastric disease
- EGUS
equine gastric ulcer syndrome
- ESGD
equine squamous gastric disease
- NSAID
nonsteroidal anti‐inflammatory drug
- OTU
operational taxonomical unit
- PCoA
principal coordinate analysis
- PPI
proton pump inhibitor
1. INTRODUCTION
Equine gastric ulcer syndrome (EGUS) is common in horses and is associated with abdominal pain, loss of appetite, poor body condition, and decreased performance.1, 2 Equine glandular gastric disease (EGGD) is described separately from equine squamous gastric disease (ESGD) as it is recognized that ulcers of each location likely differ in pathogenesis.1 Equine glandular gastric disease is a common disease across many breeds and disciplines, but particularly among horses actively in training or competition.3, 4, 5, 6, 7, 8, 9, 10 Despite the large number of horses affected by EGGD, the pathogenesis of this disease remains relatively unknown.
The pathogenesis of ESGD appears to be secondary to increased exposure to acidic gastric contents.11, 12 However, this is unlikely the primary mechanism for development of EGGD because of the protective mechanisms of the glandular mucosa and the poorer response of EGGD to proton‐pump inhibitor (PPI) therapy.13 Other potential factors that have been proposed include the roles of decreased protective factors (prostaglandins), nonsteroidal anti‐inflammatory drug (NSAID) administration, decreased gastric blood supply, and increased stress.14, 15, 16, 17 In humans, changes in the gastric microbiome have been associated with gastritis, but the gastric microbiome of horses in association with gastric glandular disease remains relatively unexplored.18, 19, 20 A prior study examining the gastric microbiome in a small number of horses (n = 10) identified differences in the glandular mucosal microbiome between horses with and without EGUS.18 However, that study included both EGGD and ESGD affected horses, and did not specify whether the glandular biopsy obtained from each horse was from normal or affected mucosa. Furthermore, management practices among the horses varied, likely leading to substantial variability in the gastric microbiome among individuals.18, 21, 22 Whether the findings from that study could be extrapolated to horses with EGGD remains to be determined.
The specific objectives of this study were to investigate the potential association between changes in the gastric microbiome and EGGD by: (a) comparing the microbiome of the gastric fluid and the glandular gastric mucosa in horses with and without EGGD and (b) comparing the glandular gastric mucosal microbiome from apparently normal and affected mucosa in horses with and without EGGD. Our hypothesis was that there would be differences in the gastric microbiome associated with the presence of EGGD.
2. MATERIALS AND METHODS
2.1. Horses and sample collection
This was a prospective, case control study that adhered to the animal handling protocol as approved by the Louisiana State University Institutional Animal Care and Use Committee (IACUC protocol number: 18‐052). Twenty‐four horses were enrolled from the institution's research herd. Horses were kept on the same pasture supplemented with grass hay and pelleted feed (Nutrena SafeChoice, Cargill Incorporated, Minneapolis, Minnesota) with no medications for at least 4 weeks prior to sample collection. The day before gastroscopy, horses were brought into a stall from the pasture and fasted for 14‐18 hours prior to the procedure. Horses were sedated with 0.4 mg/kg IV xylazine (XylaMed, Bimeda, Carrickmines, Dublin, Ireland) and gastric endoscopy performed using a 3 m gastroscope (Karl Storz SE and Co. KG, Tuttlingen, Germany). Glandular and squamous mucosa disease was evaluated as described by the European College of Equine Internal Medicine1 and a severity score assigned using the semiquantitative scale initially published by Andrews et al and modified by Sykes et al.23, 24 Gastric fluid and biopsy samples were collected from 6 horses without EGGD (controls; EGGD = 0), 8 horses with hyperemia (EGGD = 1), and 10 horses with EGGD ≥2 for a total of 24 horses. As the etiology of hyperemic lesions (with intact mucosa) versus EGGD≥2 (characterized by disrupted mucosa) may differ, samples were categorized as EGGD 1 or EGGD ≥2.
After aspiration of 20 mL gastric fluid via the biopsy channel of the endoscope, an additional 4 mL of gastric fluid was collected using a sterile syringe and flash frozen in liquid nitrogen. Endoscopic biopsy forceps (2.3 mm oval cupped with spike) were used for mucosal sample collection (Karl Storz SE and Co. KG, Tuttlingen, Germany). Three mucosal biopsies were collected of normal appearing glandular mucosa from each horse. In affected horses (EGGD >0), an additional 3 biopsies were collected from affected areas. Biopsies were rinsed with sterile saline, placed in a cryotube, then flash frozen in liquid nitrogen. Fluid and biopsy samples were then stored at −80°C until analysis. The biopsy forceps were disinfected with (dipped in) bleach and rinsed with sterile water between collections of different endoscopic appearances. The endoscope biopsy channel was disinfected (Rescue, Virox Animal Health, Oakville, ON, Canada) according to labeled directions (10 minutes contact time), and rinsed with approximately 100 mL of sterile water between horses.
2.2. Sample processing
DNA was extracted using a commercial kit (QIAamp PowerFecal DNA Kit, Qiagen, Hilden, Germany) from gastric fluid and gastric biopsies, according to the manufacturer's recommendations, with minor adaptations as previously described.21 16S rRNA amplicon library construction and sequencing was performed at the University of Missouri DNA Core facility. Concentrations of DNA for each sample were determined fluorometrically (Qubit 2.0, Invitrogen, Carlsbad, California) using quant‐iT BR dsDNA reagent kits (Invitrogen) and all samples normalized to a standard concentration for PCR amplification. Bacterial 16S rRNA amplicons were generated via amplification of the V4 hypervariable region of the 16S rRNA gene using single‐indexed universal primers (U515F/806R) flanked by Illumina standard adapter sequences and the following parameters: 98°C(3:00) + [98°C(0:15) + 50°C(0:30) + 72°C(0:30)] × 25 cycles +72°C(7:00). Amplicons were then pooled for sequencing using the Illumina MiSeq platform and V2 chemistry with 2 × 250 bp paired‐end reads, as previously described.25
All informatics analysis was performed at the MU Informatics Research Core Facility. Primers designed to match the 5′ ends of forward and reverse reads were removed from the forward read using Cutadapt.26 Reverse complements of the primer to any reverse reads present were removed from the forward read. For reverse reads, a similar, but opposite approach was performed. Read pairs were rejected if either did not match a 5′ primer with an allowable error rate of 0.1. To denoise, dereplicate, and count amplicon sequence variants (ASVs), the QIIME227 DADA2 plugin28 (version 1.10.0) was used with R version 3.5.1 and Biom version 2.1.7. The Silva.v13229 database was used to assign final taxonomies.
2.3. Statistical analysis
The signalment of horses were compared among EGGD score groups using Kruskal Wallis (age) and Fisher's exact (breed and sex) tests. Breed was dichotomized to Thoroughbred and non‐Thoroughbred. The proportion of samples that met the inclusion criteria of greater than 1000 read counts were compared among EGGD score groups and endoscopic appearance groups by Chi square with Bonferroni correction using GraphPad Prism 8 (GraphPad, San Diego, California). For further analysis, only biopsies or gastric fluid samples with >1000 read counts were included. Relative abundance data from repetitive samples that met the inclusion criteria were averaged using Microsoft Excel (Microsoft, Redmond, Washington). Samples were categorized by 2 grouping methods: (a) by EGGD score: 0 (EGGD 0‐BX), 1 (EGGD 1‐BX), and ≥ 2 (EGGD ≥2‐BX); and (b) by endoscopic appearance: controls (EGGD 0‐BX) from horses with EGGD score 0, normals (NORM‐BX) from endoscopically normal mucosa in horses with EGGD (score ≥ 1), hyperemic areas (HYPER‐BX) of intact mucosa in horses with EGGD, and areas of disrupted mucosa identified as lesions (LESION‐BX) in horses with EGGD score ≥ 2. When grouped by score, biopsies from each horse were averaged regardless of endoscopic appearance. Relative abundance data at the phylum, genus, and species level were compared across groups via Kruskal‐Wallis using IBM SPSS Statistics Version 25 (IBM Corporation, Armonk, New York) followed by Benjamini Hochberg procedure for a 25% false discovery rate. To identify overall compositional differences, similarity indices of rarefied ASVs and the relative abundance data at the L6 (genus) level were analyzed by 1‐way permutational multivariate analysis of variance (PERMANOVA) using 9999 permutations with Bonferroni correction of pairwise comparisons in Past 4.04 software package (University of Oslo, Oslo, Norway).30 Jaccard and Bray‐Curtis similarity indices were used to assess for compositional differences based on what taxa were present as well as for the effect of taxa present in combination with the relative abundance of those taxa. PERMANOVA results were reported using the F statistic for comparison of group means (greater F statistic indicates greater difference). Principal coordinate analysis (PCoA) was performed on ¼ root‐transformed relative abundance data using Past 4.04. Diversity indices of ASVs were calculated in Past 4.04 then analyzed via t‐test using GraphPad. Significance was set at P‐value ≤.05.
3. RESULTS
Twenty‐four horses were enrolled in this study: 6 horses with score 0, 8 with score 1, 9 with score 2, and 1 with score 3. The lesion phenotypes found in horses with score ≥ 2 were flat hemorrhagic (n = 3), flat hemorrhagic + flat fibrinosuppurative (n = 2), flat hemorrhagic + flat fibrinosuppurative + raised fibrinosuppurative (n = 1), flat fibrinosuppurative + raised hemorrhagic (n = 1), flat fibrinosuppurative (n = 1), raised fibrinosuppurative (n = 1), and raised hemorrhagic (n = 1). Because of the small numbers, all phenotypes were considered together (LESION‐BX). The mean age was 10.9 years (±4.1 years). Breeds represented included: Thoroughbred (n = 19), Quarter Horse (n = 3), Warmblood (n = 1), and Arabian (n = 1). There were 10 mares and 14 geldings. There were no differences detected in age (P = .56), breed (P = .35), or sex (P = .45) among groups. A total of 135 biopsies were taken with distributions among groups as listed in Tables 1 and 2. A greater proportion of biopsies from horses with EGGD ≥2 met inclusion criteria (>1000 read counts) compared to biopsies from horses with EGGD 0 (P = .05). Additionally, a greater proportion of LESION‐BX met inclusion criteria (>1000 read counts) compared to NORM‐BX (P = .04). Ninety‐one biopsies (EGGD 0, n = 9; EGGD 1, n = 32; EGGD ≥2, n = 50) met the inclusion criteria of >1000 read counts for further analysis (Tables 1 and 2). The median number of biopsies included per horse without EGGD was 1.5 (range, 0‐2). For horses with EGGD score 1 there was a median of 4.5 biopsies per horse (range, 1‐6) and for horses with EGGD score ≥2 median of 4.5 (range, 3‐8) (Supplementary Table 1). These replicates were averaged per horse for comparison among EGGD score groups and by endoscopic appearance of the biopsy site for each horse for comparison among endoscopic appearance groups. For gastric fluid samples, 10/24 samples (EGGD 0, n = 4; EGGD 1, n = 3; EGGD ≥2, n = 3) met the inclusion criteria of >1000 reads (Table 1).
TABLE 1.
EGGD score distribution of horses enrolled and number of biopsies collected from each group
| EGGD score | Number of horses | Number of biopsies | Number of horses with at least one biopsy sample included | Number of biopsies with >1000 reads |
|---|---|---|---|---|
|
0a (EGGD 0‐BX) |
6 | 18 | 5 | 9 |
|
1 (EGGD 1‐BX) |
8 | 45 | 8 | 32 |
|
≥2b (EGGD ≥2‐BX) |
10 | 72 | 10 | 50 |
| Totals | 24 | 135 | 23 | 91 |
Note: Significant difference in proportion of samples that met inclusion criteria between groups with different letter superscript.
TABLE 2.
Number of samples based on endoscopic appearance of mucosa at the biopsy site
| Endoscopic appearance | Number of biopsies | Number of biopsies with >1000 reads |
|---|---|---|
|
Control (EGGD 0‐BX) |
18 | 9 |
|
Grossly normala (NORM‐BX) |
54 | 29 |
|
Hyperemic (HYPER‐BX) |
34 | 24 |
|
Lesionb (LESION‐BX) |
29 | 25 |
| Totals | 135 | 87 |
Note: Significant difference in proportion of samples that met inclusion criteria between groups with different letter superscript.
3.1. Major taxa detected
There were 33 phyla, 960 genera, and 1537 species detected. The major phyla for the mucosal biopsy samples were Proteobacteria (50% average relative abundance), Bacteroidetes (21%), and Firmicutes (20%) (Figure 1). The major genera detected in the biopsies were Actinobacillus (17% average relative abundance), Moraxella (10%), and Porphyromonas (9%) (Figure 2). For the gastric fluid microbiome samples, the major phyla were Proteobacteria (39% average relative abundance), Firmicutes (28%), and Bacteroidetes (19%) and the major genera were Actinobacillus (13%), Lactobacillus (8%), and Alloprevotella (7%). Helicobacter species were detected, but not in association with EGGD. There was no significant difference detected in relative abundance of any individual taxa among score or biopsy type groups for the phylum, genus, and species levels.
FIGURE 1.
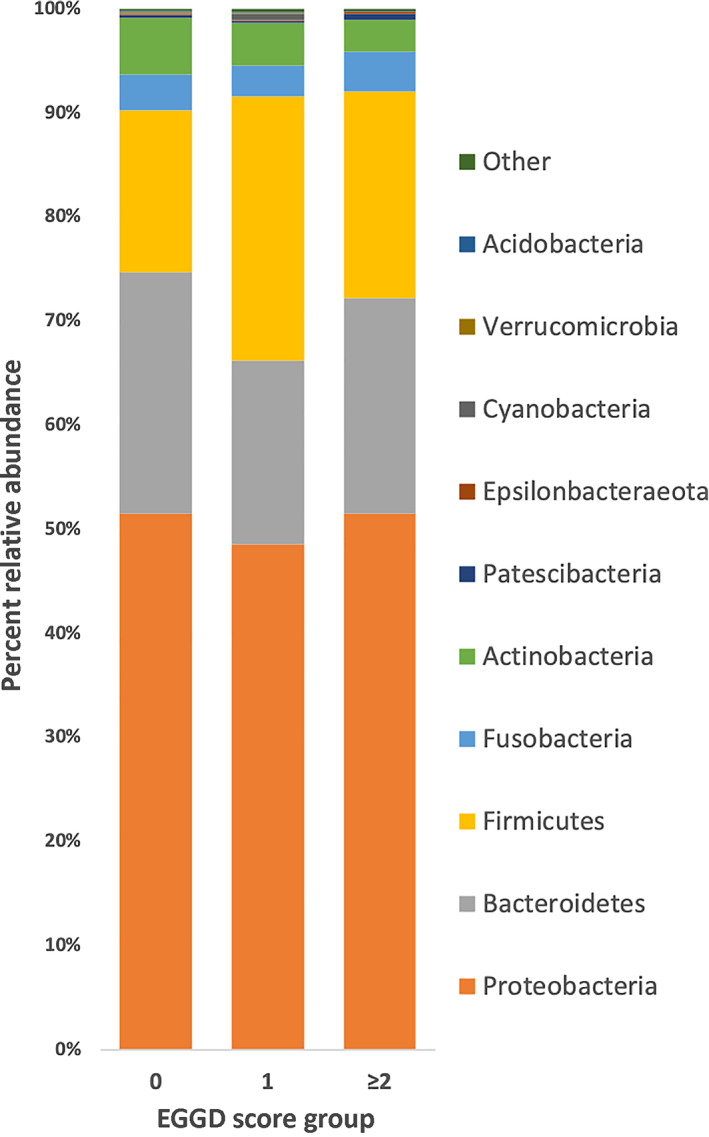
Stacked bar chart depicting the major phyla detected in gastric mucosa biopsy samples from horses without EGGD (EGGD 0), with EGGD 1, and with EGGD ≥2
FIGURE 2.
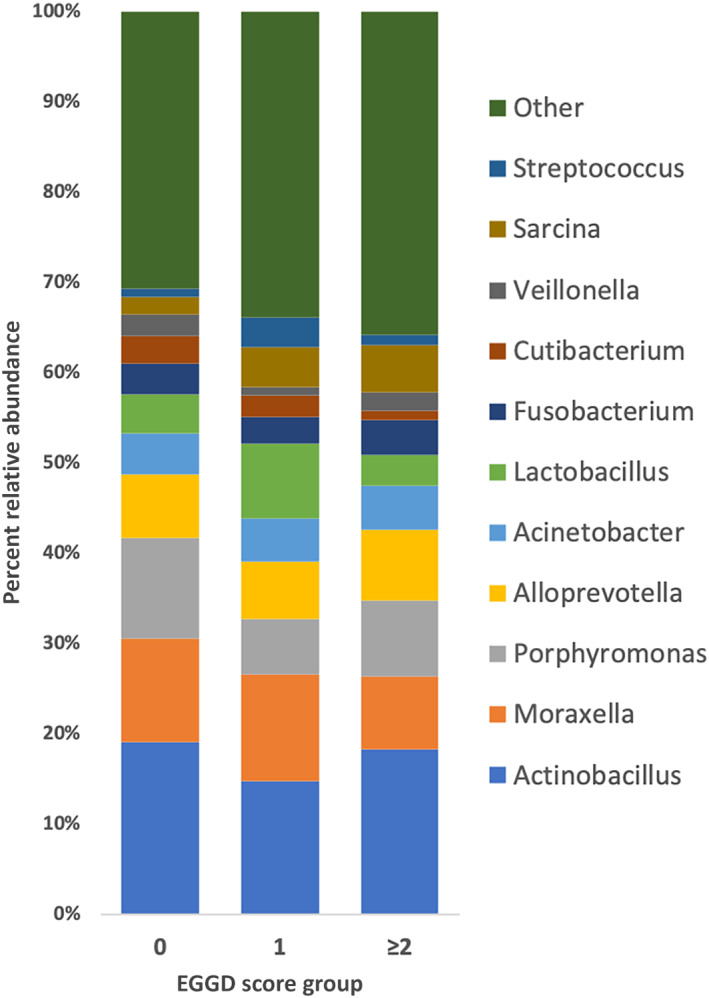
Stacked bar chart depicting the major genera detected in gastric mucosa biopsy samples from horses without EGGD (EGGD 0), with EGGD 1, and with EGGD ≥2
3.2. Comparisons of mucosal microbiomes
There were modest differences in the community structure of the gastric glandular mucosal microbiome among EGGD score groups when analyzed at the L6 (genus) level with the Jaccard similarity index (P = .009; F = 1.51). EGGD 0‐BX differed from EGGD ≥2‐BX (P = .0006). Principal coordinate analysis of EGGD score groups at L6 with Jaccard and Bray‐Curtis similarity indices showed separate clusters of EGGD 0‐BX and EGGD ≥2‐BX (Figure 3). A difference in ASV community structure was also detected when comparing EGGD 0‐BX and EGGD ≥2‐BX (Jaccard similarity index, P = .05; F = 1.34). There was no difference detected among EGGD score groups with the Bray‐Curtis similarity index (P = .98; F = 0.55). Comparisons among endoscopic appearance groups (EGGD 0‐BX, NORM‐BX, HYPER‐BX, LESION‐BX) for either similarity index detected no differences (Jaccard P = .6, F = 0.97; Bray‐Curtis P = .97, F = 0.66).
FIGURE 3.
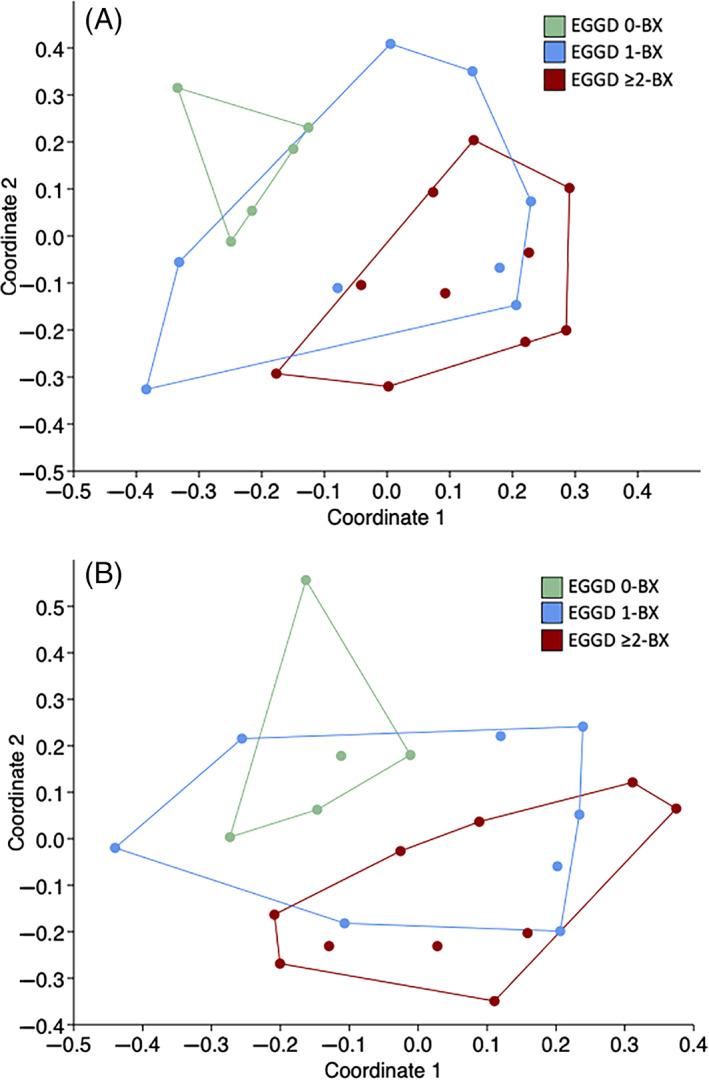
Principal coordinate analysis of gastric mucosal biopsy samples at the genus level. A, EGGD score compared using the Jaccard similarity index (P = .009). B, EGGD score compared using the Bray‐Curtis similarity index (P = .98)
There were no differences among score groups for diversity indices of ASVs: Chao1 (P = .12), Shannon H (P = .79), individuals (P = .25), and total number of taxa detected (P = .33).
3.3. Comparison of mucosal microbiome to gastric fluid microbiome
Only horses that had mucosal biopsies and gastric fluid samples that met inclusion criteria were used for comparison of these microbiomes (n = 9). There was an overall modest difference in community structure at the L6 (genus) level detected between gastric fluid samples and biopsy samples using the Jaccard similarity index (P = .005; F = 2.32). No difference was detected using the Bray‐Curtis similarity index (P = .59; F = 0.75). No differences were detected when the gastric fluid and biopsy samples were compared within EGGD score 0 and EGGD score ≥2 groups. No visual clustering patterns were observed in the PCoA models.
4. DISCUSSION
In the study presented here, there was a modest difference in the microbiome of mucosal samples that was associated with EGGD scores. There was a modest difference in the overall microbiome community structure among EGGD score groups with greatest differences detected between EGGD score 0 and score ≥2. This difference was further supported by the separate clusters of EGGD 0‐BX and EGGD ≥2‐BX observed in the PCoA model.
The findings of the present study build upon prior studies of the microbiome in EGUS18, 22 by specifically characterizing the microbiome of healthy glandular mucosa and EGGD lesions. Consistent with previous reports, Proteobacteria, Firmicutes, and Bacteroidetes were the major phyla,18, 21, 22, 31 and Moraxella, Actinobacillus, and Porphyromonas were the major genera of the glandular mucosa.21, 22, 31, 32 There were modest differences observed in the community structure of the glandular mucosal microbiome among groups in this study that might indicate a bacterial association with development or persistence of EGGD. Specifically, differences were detected among EGGD score groups and not endoscopic appearance groups suggesting that there might be global alterations in the mucosal microbiome of horses with EGGD. In people with non‐Helicobacter, non‐NSAID‐associated gastritis, there are differences in the gastric microbiome, specifically an increase in the relative abundance of Firmicutes in association with disease.19 There was no single bacterial species, phylum, or genus associated with EGGD in the present study.
In humans, peptic ulcers are strongly associated with the presence of Helicobacter pylori in the gastric mucosa.33 This association led to multiple studies investigating the role of Helicobacter spp. in EGUS.18, 22, 34, 35, 36, 37 However, to date, there has been no convincing evidence of Helicobacter as a primary cause of EGUS.18, 22, 34, 35, 36, 37 In the present study, Helicobacter spp. were detected, but not in association with EGGD. Helicobacter does not appear to be associated with EGGD in horses.
The differences found in this study were consistently detected using the Jaccard similarity index. This index is a binary test that looks at the presence or absence of sequences representing different bacterial taxa. Conversely, no differences were detected using the Bray‐Curtis index which also takes into account the relative abundance of the sequences present. While there were no differences detected for any individual taxa among groups, this data supports that the presence or absence of specific bacteria might be associated with EGGD, rather than a dysbiosis. Identification of a specific bacterial taxa associated with the formation or persistence of EGGD would allow for targeted investigation into the use of antibiotics or probiotics as part of the therapeutic plan for affected horses. Clinically, horses with persistent EGGD might be treated with antibiotics if they are nonresponsive to conventional gastroprotectant therapies.38 Previous research into antibiotic therapy for EGGD has yet to support this practice and based on antimicrobial stewardship guidelines cannot currently be recommended.39 For the treatment of peptic ulcer disease in people positive for H. pylori, antimicrobial therapy is used to eradicate these bacteria as part of the treatment protocol.40 If a specific bacterial target can be found in horses with EGGD, then intervention by manipulation of the microbiome could improve success of treatment or preventive strategies.
Limitations of this study include a small sample size, mild EGGD, and small biopsy mass. Small sample size could have limited our ability to detect differences between groups. This is compounded by the fact that the stomach has a higher variability in microbiome among horses, when compared to the hindgut,21, 31 which might be associated with different environments, diets, and management practices.18, 21, 22 To decrease these impacts on interhorse variation of the gastric microbiome, we sampled horses that were in the same environment and under the same management practices including diet. However, this could mean that differences observed in this study do not apply broadly to horses in other environments or under different management practices. An additional limitation to the study was the small biopsy size, because of dimensions of the biopsy forceps, that provided a low biomass sample. This likely contributed to the low number of read counts for some samples. The variation in number of samples among groups could have also contributed to the differences detected.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Approved by Louisiana State University IACUC, protocol number: 18‐052.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Supplementary Table 1. Number of samples included from each horse.
ACKNOWLEDGMENT
Funding provided by Louisiana State University Equine Health Studies Program Charles V. Cusimano Grant. We acknowledge Christian Arias, Amanda Distefano, Jenny Windham, and Bailey Clouatre for their assistance with sample collection.
Paul LJ, Ericsson AC, Andrews FM, et al. Gastric microbiome in horses with and without equine glandular gastric disease. J Vet Intern Med. 2021;35(5):2458‐2464. 10.1111/jvim.16241
Funding information LSU Equine Health Studies Program Charles V. Cusimano Grant
REFERENCES
- 1.Sykes B, Hewetson M, Hepburn R, et al. European College of Equine Internal Medicine Consensus Statement—Equine gastric ulcer syndrome in adult horses. J Vet Intern Med. 2015;29:1288‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sykes BW, Bowen M, Habershon‐Butcher JL, Green M, Hallowell GD. Management factors and clinical implications of glandular and squamous gastric disease in horses. J Vet Intern Med. 2019;33:233‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luthersson N, Nielsen KH, Harris P, et al. The prevalence and anatomical distribution of equine gastric ulceration syndrome (EGUS) in 201 horses in Denmark. Equine Vet J. 2009;41:619‐624. [DOI] [PubMed] [Google Scholar]
- 4.Murray MJ, Schusser GF, Pipers FS, et al. Factors associated with gastric lesions in Thoroughbred racehorses. Equine Vet J. 1996;28:368‐374. [DOI] [PubMed] [Google Scholar]
- 5.Begg LM, O'Sullivan CB. The prevalence and distribution of gastric ulceration in 345 racehorses. Aust Vet J. 2003;81:3. [DOI] [PubMed] [Google Scholar]
- 6.Habershon‐Butcher JL, Hallowell GD, Bowen IM, et al. Prevalence and risk factors for ulceration of the gastric glandular mucosa in Thoroughbred racehorses in training in the UK and Australia. Paper presented at: Research Abstracts Program ACVIM Forum; 2012; New Orleans, LA.
- 7.Banse HE, MacLeod H, Crosby C, Windeyer MC. Prevalence of and risk factors for equine glandular and squamous gastric disease in polo horses. Can Vet J. 2018;59:880‐884. [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen S, Cribb A, Windeyer M, et al. Risk factors for equine glandular and squamous gastric disease in show jumping Warmbloods. Equine Vet J. 2018;50:747‐751. [DOI] [PubMed] [Google Scholar]
- 9.Tamzali Y, Marguet C, Priymenko N, et al. Prevalence of gastric ulcer syndrome in high‐level endurance horses. Equine Vet J. 2011;43:141‐144. [DOI] [PubMed] [Google Scholar]
- 10.Nieto JE, Snyder JR, Beldomenico P, Aleman M, Kerr JW, Spier SJ. Prevalence of gastric ulcers in endurance horses—a preliminary report. Vet J. 2004;167:33‐37. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzo‐Figueras M, Merritt AM. Effects of exercise on gastric volume and pH in the proximal portion of the stomach of horses. Am J Vet Res. 2002;63:1481‐1487. [DOI] [PubMed] [Google Scholar]
- 12.Nadeau JA, Andrews FM, Patton CS, Argenzio RA, Mathew AG, Saxton AM. Effects of hydrochloric, acetic, butyric, and propionic acids on pathogenesis of ulcers in the nonglandular portion of the stomach of horses. Am J Vet Res. 2003;64:404‐412. [DOI] [PubMed] [Google Scholar]
- 13.Sykes BW, Sykes KM, Hallowell GD. A comparison of three doses of omeprazole in the treatment of equine gastric ulcer syndrome: a blinded, randomised, dose‐response clinical trial. Equine Vet J. 2015;47:285‐290. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen SK, Cribb AE, Read EK, French D, Banse HE. Phenylbutazone induces equine glandular gastric disease without decreasing prostaglandin E2 concentrations. J Vet Pharmacol Ther. 2018;41:239‐245. [DOI] [PubMed] [Google Scholar]
- 15.MacAllister CG, Morgan SJ, Borne AT, Pollet RA. Comparison of adverse effects of phenylbutazone, flunixin meglumine, and ketoprofen in horses. J Am Vet Med Assoc. 1993;202:71‐77. [PubMed] [Google Scholar]
- 16.Rendle D, Bowen M, Brazil T, et al. Recommendations for the management of equine glandular gastric disease. UK‐Vet Equine. 2018;2:2‐11. [Google Scholar]
- 17.Scheidegger MD, Gerber V, Bruckmaier RM, van der Kolk JH, Burger D, Ramseyer A. Increased adrenocortical response to adrenocorticotropic hormone (ACTH) in sport horses with equine glandular gastric disease (EGGD). Vet J. 2017;228:7‐12. [DOI] [PubMed] [Google Scholar]
- 18.Dong HJ, Ho H, Hwang H, et al. Diversity of the gastric microbiota in Thoroughbred racehorses having gastric ulcer. J Microbiol Biotechnol. 2016;26:763‐774. [DOI] [PubMed] [Google Scholar]
- 19.Li XX, Wong GL, To KF, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non‐steroidal anti‐inflammatory drug use. PLoS One. 2009;4:e7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung JJY, Coker OO, Chu E, et al. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut. 2020;69:1572‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ericsson AC, Johnson PJ, Lopes MA, Perry SC, Lanter HR. A microbiological map of the healthy equine gastrointestinal tract. PLoS One. 2016;11:e0166523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins GA, den Bakker HC, Burton AJ, et al. Equine stomachs harbor an abundant and diverse mucosal microbiota. Appl Environ Microbiol. 2012;78:2522‐2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sykes B, Jokisalo J. Rethinking equine gastric ulcer syndrome: part 1—terminology, clinical signs and diagnosis. Equine Vet Educ. 2014;26:543‐547. [Google Scholar]
- 24.Andrews FM, Bernard W, Byars D, et al. Recommendations for the diagnosis and treatment of equine gastric ulcer syndrome (EGUS): the equine gastric ulcer council. Equine Vet Educ. 1999;11:262‐272. [Google Scholar]
- 25.Ericsson AC, Davis JW, Spollen W, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One. 2015;10:e0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin M. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnetjournal. 2011;17:10‐12. [Google Scholar]
- 27.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188‐7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
- 31.Costa MC, Silva G, Ramos RV, et al. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet J. 2015;205:74‐80. [DOI] [PubMed] [Google Scholar]
- 32.Burton AB, Perkins GA, Parker J, et al. The gastric mucosa of horses harbors an abundant and diverse bacterial flora. Paper presented at: Research Abstract Program ACVIM Forum; 2007; Seattle, WA.
- 33.Dorer MS, Talarico S, Salama NR. Helicobacter pylori's unconventional role in health and disease. PLoS Pathog. 2009;5:e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Husted L, Jensen TK, Olsen SN, et al. Examination of equine glandular stomach lesions for bacteria, including Helicobacter spp by fluorescence in situ hybridisation. BMC Microbiol. 2010;10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martineau H, Thompson H, Taylor D. Pathology of gastritis and gastric ulceration in the horse. Part 1: range of lesions present in 21 mature individuals. Equine Vet J. 2009;41:638‐644. [DOI] [PubMed] [Google Scholar]
- 36.Contreras M, Morales A, Garcia‐Amado MA, et al. Detection of Helicobacter‐like DNA in the gastric mucosa of Thoroughbred horses. Lett Appl Microbiol. 2007;45:553‐557. [DOI] [PubMed] [Google Scholar]
- 37.Morales A, Garcia F, Bermudez V. Detection of Helicobacter‐like organisms in Thoroughbred horses from Venezuela. Braz J Vet Pathol. 2010;3:4. [Google Scholar]
- 38.Buchanan BR, Andrews FM. Treatment and prevention of equine gastric ulcer syndrome. Vet Clin North Am Equine Pract. 2003;19:575‐597. [DOI] [PubMed] [Google Scholar]
- 39.Sykes BW, Sykes KM, Hallowell GD. Administration of trimethoprim‐sulphadimidine does not improve healing of glandular gastric ulceration in horses receiving omeprazole: a randomised, blinded, clinical study. BMC Vet Res. 2014;10:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanas A, Chan FKL. Peptic ulcer disease. Lancet. 2017;390:613‐624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Number of samples included from each horse.


