Extended Data Fig. 9 |. GBPL3 IDR contributes to LLPS-driven defence.
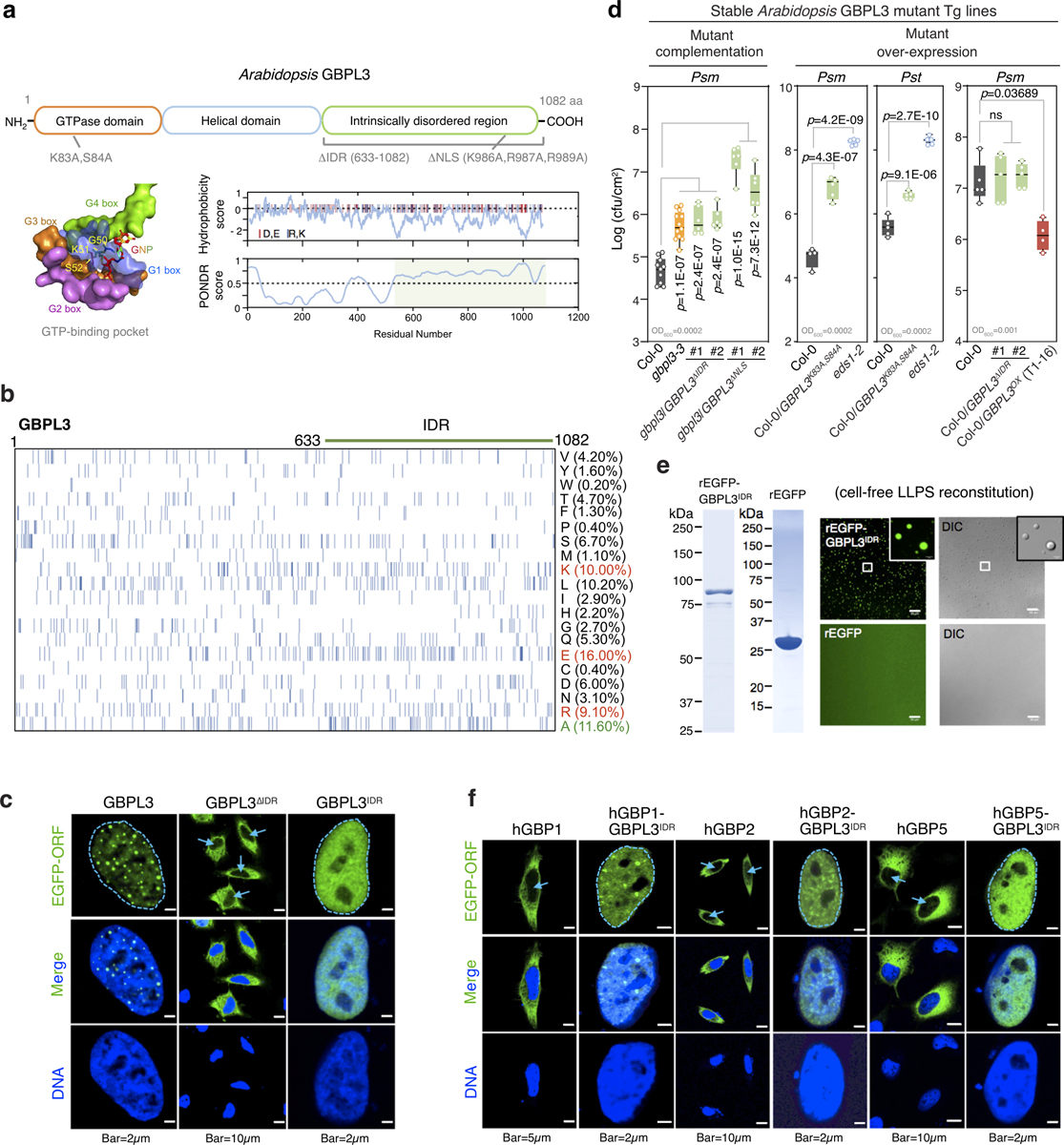
a, Top, domain structures of the Arabidopsis GBPL3 protein. Sites for mutagenesis are shown below. Bottom, GTP-binding pocket with conserved G-boxes (hGBP1 surface representation; 1F5N.PDB). GBPL3 hydrophobicity and disorder plots on right. Charged amino acids (red, glutamate and aspartate; blue, arginine and lysine) displayed as vertical lines atop panels. IDRs are shaded light green. b, Amino acid compositions of GBPL3 full length protein. Each row represents information for a single amino acid. Vertical bars, occurrence of indicated amino acid at that position. Right, amino acid percentage. c, Immunofluorescence of EGFP−GBPL3 mutant variants in HeLa cells 36 h after transfection. DNA stained with Hoechst 33342 dye. Arrows, nucleus. d, Growth of Psm ES4326 (Psm) and Pst DC3000 (Pst) at day 3 of infection. Inoculum, bottom left. Statistical analysis, comparison of mean via one-way ANOVA test (Bonferroni post hoc correction). ns, not significant. Box = 25th and 75th percentiles; bars = min and max values. Individual data points represent biologically independent samples. e, In vitro phase separation of rEGFP−GBPL3IDR and rEGFP. Equimolar (400 nM) amounts incubated at 22 °C for 16 h in droplet buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP, 10% Ficoll). Bar, 20 μm. Left, Coomassie stain. f, Immunofluorescence of plant−human GBP-IDR chimeras performed as in c.
