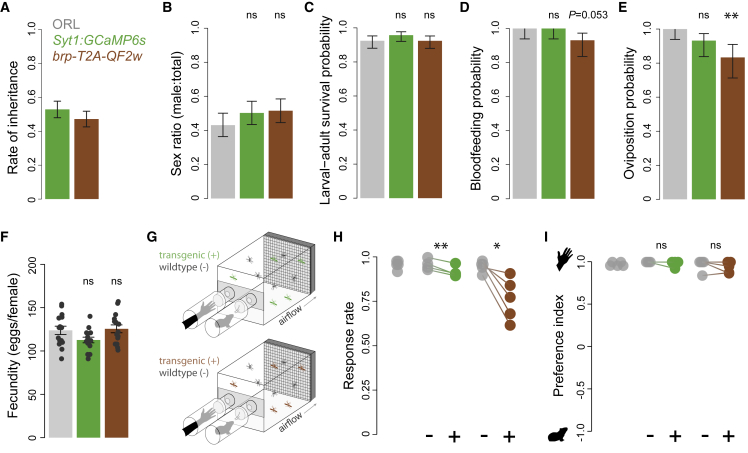
Figure 6.
Life history and olfactory behavior of pan-neuronal strains
Syt1:GCaMP6s (green) and brp-T2A-QF2w (brown, no effector present) heterozygotes were tested alongside wild-type Orlando (gray) mosquitoes.
(A–F) (A) Transgenic constructs were inherited by ~50% of offspring in heterozygote × wild-type crosses. (B–F) Transgenic lines resembled wild type in key life history traits (A–F), except that brp-T2A-QF2w females showed modest reductions in blood-feeding and oviposition rates. Bars and lines in (A) to (E) indicate point estimates and 95% confidence intervals of binomial probabilities for each trait (n = 397–445 [A], 194–210 [B and C], 54–59 [D and E] females per genotype). Bars and lines in (F) indicate mean ± SEM for n = 15 females. Statistical significance between each transgenic genotype and wild type was assessed by using chi-squared tests of independence (B–E) and t tests (F). Note that chi-squared tests for (B) to (E) agreed with comparisons of binomial confidence intervals.
(G) Schematic of two-port olfactometer assays used to test preference for human versus guinea pig odor. Transgenic heterozygotes were tested alongside their wild-type siblings in a paired design. The wild-type ORL strain was also tested separately (not depicted).
(H and I) Response rate (H) and preference index (I) from preference trials. Lines connect data points for sibling groups from the same trial, and significance was assessed by using paired t tests
∗p < 0.05; ∗∗p < 0.01; ns, not significant.