Figure 2.
Endogenous 5hmC-based lineage reconstruction using scPECLR
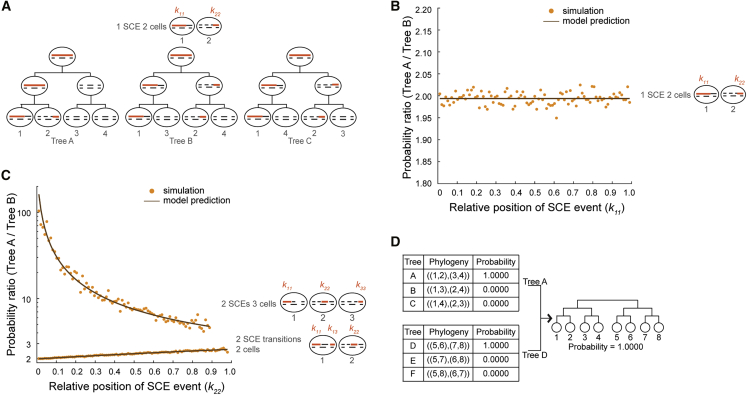
(A) Two cells sharing an original DNA strand (solid orange line) can either be sisters (Tree A) or cousins (trees B and C) depending on whether the SCE event occurred at the 4- to 8-cell or 2- to 4-cell stage, respectively. Newly synthesized DNA strands are shown as dashed black lines.
(B) For an SCE transition between two cells, the probability of the pair of cells being sisters versus cousins is plotted against the relative position of the SCE event on the chromosome (). The model prediction (black) and simulation results (yellow) are shown for chromosome 1 (N = 97 for 2-Mb bins) with
(C) The probability ratio between Trees A and B are shown for N = 97 and for two cases: two SCE transitions shared between two cells and two SCE events shared between three cells.
(D) For the 8-cell mouse embryo in Figure 1B, the probability of observing the different topologies, rounded to four decimal places, for the two 4-cell subtrees are shown.