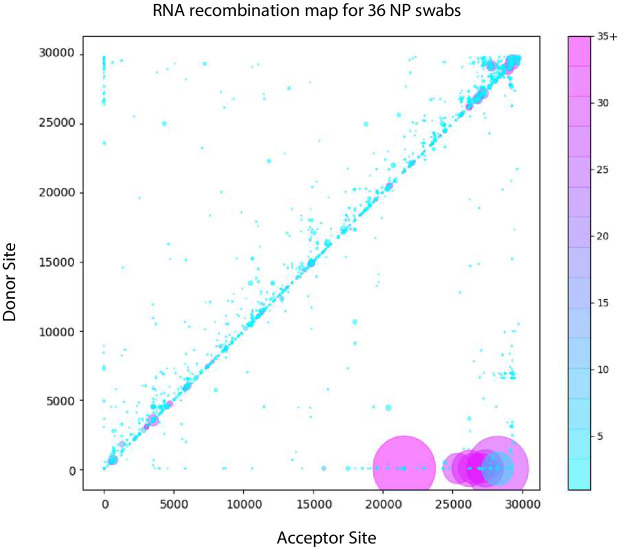
Figure 7. Tiled-ClickSeq identifies sub-genomic mRNAs, structural variants and Defective-RNAs in clinical samples of SARS-CoV-2.
Similarly to Figure 5B, unique RNA recombination events are plotted for 36 clinical samples as a scatterplot whereby the upstream ‘donor’ site is plotted on the y-axis and a downstream ‘acceptor’ site is plotted on x-axis using the WA-1 reference coordinates for each sample. The read count for each unique RNA recombination event is indicated by the size of the point, while the number of samples in which this each RNA recombination event is found is indicated by the colour-bar. Insertions/duplication/back-splicing events are found above the x = y axis, while deletions and RNA recombination events yielding sgmRNAs are found below.