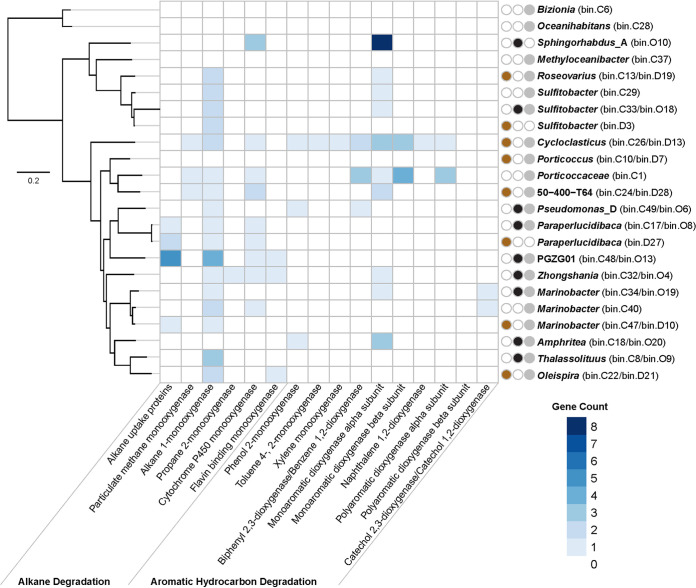
FIG 7.
Heatmap representing the gene count of select biodegradation enzymes from MAGs recovered from metagenomic libraries constructed from microcosms amended with diesel (brown) or crude oil (black) and following coassembly of the two (gray). Bin names corresponding to these MAGs are indicated on the right (labeled D, O, and C, respectively) and are arranged according to a maximum-likelihood phylogenomic tree (shown at left) based on 43 concatenated marker genes. The scale bar indicates 0.2 substitutions per nucleotide position. Enzymes containing multiple subunits were counted once if all subunits were present (see Data Set S1). Unpaired mono- or polycyclic aromatic dioxygenase alpha and beta subunits were also counted.