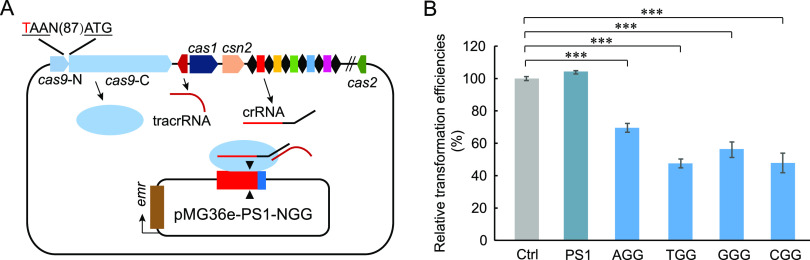
FIG 1.
Function validation of the endogenous CRISPR-Cas system in P. acidilactici LA412. (A) Schematic of the endogenous CRISPR-based genome editing in P. acidilactici LA412. On the upper part of schematic are the CRISPR array, tracrRNA gene, and cas genes (cas1, csn2, cas2, and disrupted cas9, including cas9-N and cas9-C). N(87) represents 87 bp between the ATG and TAA codons. The challenging plasmids with the cassette of “protospacer-PAM” were transformed into the cells and cleaved by the endogenous CRISPR-Cas system. Red, blue, and brown bars on the challenging plasmid indicated the protospacer which matched with spacer 1 (PS1) of the CRISPR array, the NGG PAM (in the sequence, N corresponds to A, T, G, or C), and erythromycin resistance gene emr, respectively. (B) Relative transformation efficiencies of the challenging plasmids with or without NGG PAM. The data are expressed as mean ± SD with three technical replicates. ***, P < 0.001.