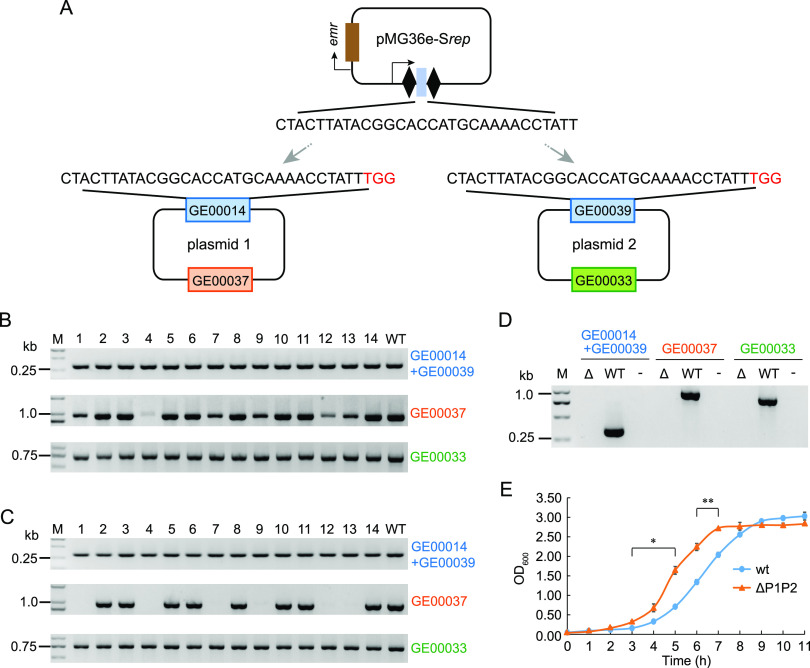
FIG 3.
Endogenous CRISPR-mediated depletion of native plasmids increased cell growth. (A) Schematic of the interference plasmid pMG36e-Srep, which simultaneously targets the replication protein genes on both native plasmid 1 (P1) and plasmid 2 (P2). These replication protein genes (GE00014 and GE00039) share one identical protospacer to be targeted. (B) Colony PCR amplification of the target gene, GE00037 gene on P1, and GE00033 gene on P2 from 14 randomly selected transformants (lanes 1 to 14). M, DNA ladder; WT, control group with P. acidilactici total DNA as the template. (C) PCR amplification of target gene, GE00037, and GE00033 from 14 transformants (lanes 1 to 14 in panel B) after one passage in modified MRS medium containing 5 μg/ml erythromycin. (D) PCR amplification of the above 3 genes from the native plasmid-depleted single colony. Δ, WT, and “−” indicate the native plasmid-depleted strain, wild-type strain, and double-distilled water (ddH2O) as the template. (E) Growth curves of wild-type (wt) and the native plasmid-depleted strain (ΔP1P2). Data are expressed as the mean ± SD with three technical replicates. *, P < 0.05; **, P < 0.01.