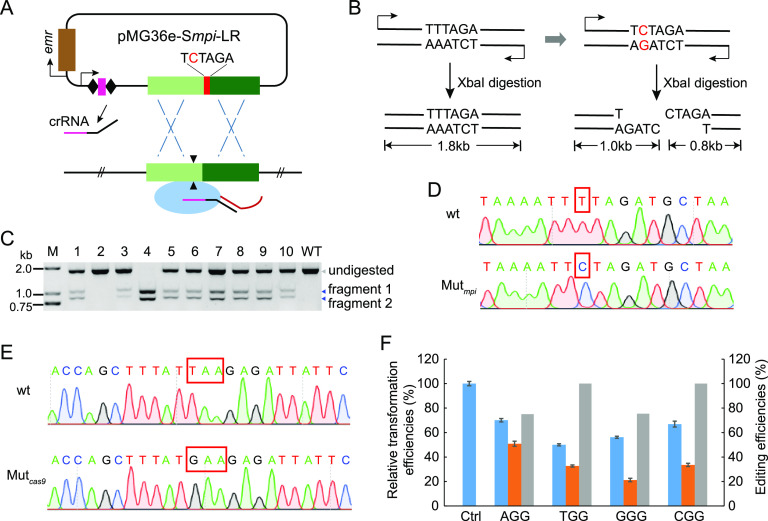
FIG 4.
Introduction of point mutation into the P. acidilactici genome. (A) Schematic of point mutation introduction based on endogenous CRISPR-Cas system. The editing plasmid cleaved the target region matching with the spacer, further inducing crossover between the target gene and the donor DNA with a desired nucleotide. (B) Schematic of point mutation detection. PCR amplification product of the wild-type mpi gene in LA412 was not digested by XbaI restriction endonuclease. However, the PCR amplification product of the mpi gene with an introduced point mutation was digested by XbaI to obtain two DNA fragments. (C) PCR products of the mpi gene amplified from 10 randomly selected transformants (lanes 1 to 10) carrying the editing plasmid or from the wild-type strain (wt) were digested by XbaI and analyzed by agarose gel. M, DNA ladder; WT, control group using mpi gene of wild-type strain as the amplification template; undigested, bands of undigested PCR products; fragments 1 and 2, digested bands. (D) Sequencing peaks of the PCR products amplified from the wild-type and point-mutated mpi gene loci. The red rectangle marks the mutation site. (E) Sequencing peaks of the PCR products amplified from the wild type (disrupted) and the corrected cas9 gene locus. The red rectangle indicates TAA stop codon and GAA codon. (F) Blue columns indicate relative transformation efficiencies of the interference plasmids targeting pyrE gene, orange columns indicate those of corresponding editing plasmids, and gray columns denote pyrE deletion efficiencies of the editing plasmids in P. acidilactici LA412cas9 strain. Data are expressed as the mean ± SD with three technical replicates.