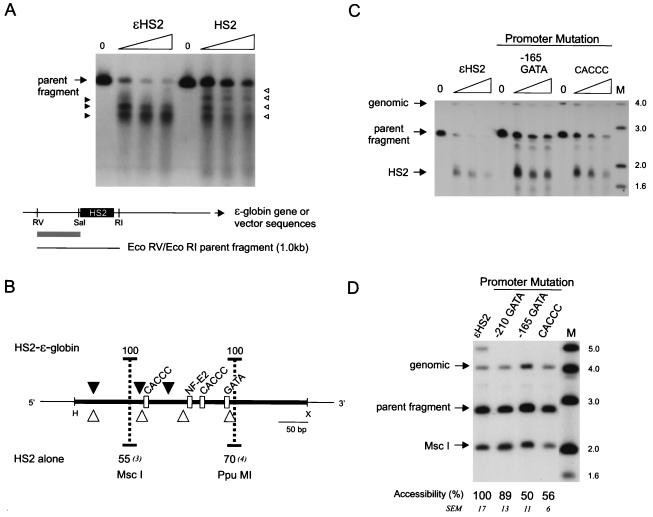
FIG. 5.
Nuclease sensitivity of HS2 in the absence of a linked globin gene and effects of promoter mutations on HS2. (A) Nuclei of K562 clones containing HS2 alone on minichromosomes were digested with DNase I as detailed in the legend to Fig. 2A, and the cleavage sites were mapped after double digestion with EcoRI and EcoRV. (B) Summary of DNase I cleavage sites in wild-type ɛHS2 and in HS2 alone. The percent accessibility and SEM at the HS2 MscI and PpuMI sites for six clones of each type are shown. The probe fragment used for both experiments was an EcoRV-to-SalI fragment flanking HS2 (gray bar). RV, EcoRV; RI, EcoRI; S, SalI. (C) Samples were processed as detailed in the legend to Fig. 2A, and the HS2 site was mapped after digestion with EcoRV. (D) Samples were processed as detailed in the legend to Fig. 2B. For the positions of DNase I and MscI cleavage within the EcoRV parent band, see Fig. 2C. Lanes M, markers with sizes given in kilobases at the right. The probe fragment used for both experiments was as detailed in the legend to Fig. 2.