Keywords: dystroglycan, post-translational glycosylation, M3 core structure, laminin-binding glycoepitope, glycosyltransferases, protein evolution
Abstract
The dystroglycan (DG) complex plays a pivotal role for the stabilization of muscles in Metazoa. It is formed by two subunits, extracellular α-DG and transmembrane β-DG, originating from a unique precursor via a complex post-translational maturation process. The α-DG subunit is extensively glycosylated in sequential steps by several specific enzymes and employs such glycan scaffold to tightly bind basement membrane molecules. Mutations of several of these enzymes cause an alteration of the carbohydrate structure of α-DG, resulting in severe neuromuscular disorders collectively named dystroglycanopathies. Given the fundamental role played by DG in muscle stability, it is biochemically and clinically relevant to investigate these post-translational modifying enzymes from an evolutionary perspective. A first phylogenetic history of the thirteen enzymes involved in the fabrication of the so-called ‘M3 core’ laminin-binding epitope has been traced by an overall sequence comparison approach, and interesting details on the primordial enzyme set have emerged, as well as substantial conservation in Metazoa. The optimization along with the evolution of a well-conserved enzymatic set responsible for the glycosylation of α-DG indicate the importance of the glycosylation shell in modulating the connection between sarcolemma and surrounding basement membranes to increase skeletal muscle stability, and eventually support movement and locomotion.
1. Background
Dystroglycan (DG) is an extracellular matrix (ECM) protein complex with a wide tissue distribution, that ranges from skeletal, cardiac and smooth muscle to the central and peripheral nervous systems [1,2]. DG is encoded by the DAG1 gene and translated from a single mRNA as a precursor which undergoes post-translational modifications that include extensive decoration with carbohydrates and processing into two subunits: the highly glycosylated extracellular α-DG and the transmembrane β-DG [3]. The two subunits interact non-covalently to form a bridge between the ECM and the actin cytoskeleton. α-DG acts as a receptor for ECM proteins containing laminin-globular (LG) domains such as laminin and agrin, among others [4]. In previous bioinformatic analyses, we have demonstrated the presence of the DG complex and gene in all the Metazoan lineages starting from Porifera, underlining its considerable biological and pathophysiological importance [5,6].
The carbohydrate moieties of α-DG are mainly concentrated within the elongated central mucin-like region that separates two-terminal globular domains [7]. Alterations in the glycosylation shell of α-DG (i.e. hypoglycosylation) can induce muscular dystrophies, referred to as primary or secondary dystroglycanopathies (hereinafter DGpathies), which present themselves with phenotypes that range from minor and later-onset to congenital and severe [8]. In the so-called primary DGpathies, caused by mutations in DAG1, α-DG glycosylation and/or the overall stability of the DG polypeptide can be affected [9]. The significance of DG integrity is stressed by the evidence that DAG1 knockout in mice is lethal during gestation, as early as day E6.5 [10], while a mutation that abolishes the entire complex and leads to post-natal mortality was found in a human family [11].
Glycosylation is one of the most important and ubiquitous forms of post-translational modification and can potentially affect the correct targeting, trafficking and sorting of a protein, thus influencing its stability and function [12]. Therefore, it is probably not a mere coincidence that the series of neuromuscular diseases collectively defined as secondary DGpathies are caused by mutations in genes coding for proteins involved in the O-glycosylation of α-DG [8,9]. Indeed, α-DG is extensively glycosylated with N-linked as well as O-linked groups [13]. O-glycosylation of α-DG follows one of the multiple O-mannosylation pathways found in eukaryotes [14]. Namely, it features the modification of some specific Thr residues, like for example Thr 317 and 319 [15–17] within the central mucin-like region of α-DG [5,7].
The α-DG core protein is heterogeneously glycosylated, and this is particularly evident in its skeletal muscle isoform [13]. In fact, the presence of a mixed population of sugars causes a characteristic ‘blurred’ appearance of the α-DG band in Western blots, indicating a sort of Gaussian distribution of molecular masses that in skeletal muscle is typically centred around 156 kDa [13]. Accordingly, a series of different ‘core carbohydrate structures' have been detected, namely the M1, M2 and M3 core structures [17]. The M3 core structure starts to be formed readily in the endoplasmic reticulum (ER) and subsequently elongated in the Golgi apparatus, while M1 and M2 are generated in the cis-Golgi, upon transport from the ER, by the action of POMGnT1 and other enzymes [18].
The heterogeneous distribution of molecular masses observed in α-DG might depend on variability in (i) the number of M1/M2 or (ii) of M3 core structures for each α-DG molecule, respectively, or in (iii) the length of each M3 core structure, and/or on differential contributions arising from each of the aforementioned aspects.
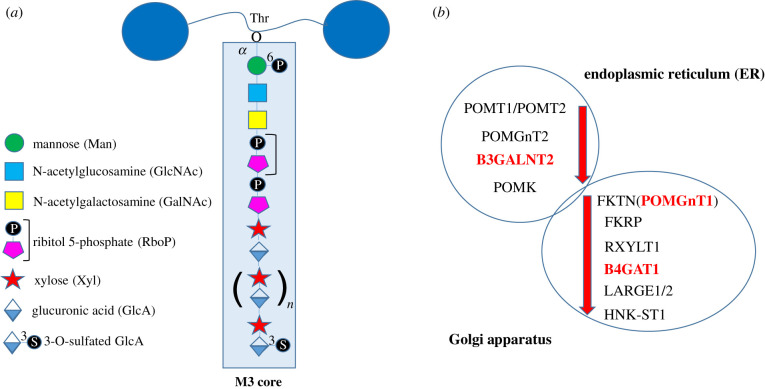
The M3 core structures are the most crucial as they include the specific glycoepitope(s) recognized by extracellular binding partners within the basement membranes surrounding skeletal muscle and other tissues [3].
Each mature α-DG molecule carries numerous (probably 20 to 30) M1 and M2 core structures and several of the Ser/Thr residues within the central mucin-like domain can be found modified with these simpler structures [17,19]. Conversely, the more complex M3 core structures represent just a minority and are found attached to as few as a couple of Thr residues in each α-DG molecule, as suggested for Thr 317 and 319 [16] or Thr 317 and 379 [17], which are highly conserved in vertebrates [5,16]. In mammals, a consensus sequence for O-mannosylation (IXPT(P/X)TXPXXXXPTX(T/X)XX) has been identified via extensive mass spectrometry analysis and found exclusively in α-DG [15]. The M3 core structure includes a long tandemly repeated polymer of several disaccharide units (xylose-glucuronic acid), defined as matriglycan [20] (figure 1a). The length of matriglycan can be variable and depends on the differential activity of LARGE1 (i.e. the enzyme directly responsible for its elongation) and HNK-1ST, which mediates sulfation at the 3-hydroxyl of the terminal glucuronic acid, thus controlling the extent and length of the overall reaction. Such competing action of the two enzymes eventually influences the resulting affinity towards laminin and other α-DG binding partners in different tissues [21]. A further complexity in the control of matriglycan synthesis emerged with the recent finding that also the kinase POMK is needed to regulate the LARGE1-mediated elongation of matriglycan [22].
Figure 1.
A scheme of the M3 core carbohydrate chain and its specific carbohydrate blocks (a) and of the progressive enzymatic cascades present in the ER and Golgi apparatus (b). All the relevant details on the consecutive enzymatic steps are reported in table 1. Reported in red in (b) are the three enzymes whose orthologues are present in all the lineages analysed.
A loss, or a reduction, of the O-mannose-bound matriglycan on α-DG can lead to a diminished cell adhesion to the ECM, with pathological consequences. Indeed, the matriglycan scaffold has a crucial physiological value since it represents the site recognized with high affinity by α-DG-binding partners [4]. The degree of overall glycosylation can vary in different tissues and might influence the affinity of α-DG towards its binding partners in a subtle and not yet fully understood way [23].
O-glycosylation is a highly controlled process that takes place in the ER/Golgi compartments and mutations in enzymes, such as TRAPPC11 and GOSR2, that are involved in localizing proteins to the Golgi compartment, have been also linked to hypoglycosylation of α-DG and muscular dystrophies [24]. In cells defective for subunits of the conserved oligomeric Golgi (COG) complex, which ‘supervises’ the proper glycosylation of proteins, an impaired glycosylation of α-DG and a consequent susceptibility to proteases [25] have been observed, reminiscent of what reported for the ‘naked’ mucin-like domain expressed in bacterial cells [26].
Aside from these general chaperones, fabricating the core M3 glycoepitope of α-DG requires a large and specific orchestra of enzymatic players, localized in the ER first and then in the cis- and median-Golgi compartments (figure 1b). A great deal of outstanding work conducted in different laboratories has revealed that in mammals there are (at least) 13 enzymes responsible for the consecutive steps leading to the mature M3 core structure [27–30]. Such a high degree of complexity highlights the importance of understanding the molecular mechanism of α-DG glycosylation for potential therapeutic applications [31].
The enzyme full names, acronyms and reaction details are reported in table 1 (see also figure 1). The next three sections describe the sequential glycosylation steps and some additional details, including the relevant human pathologies related to these enzymes that underline their functional importance in mammals.
Table 1.
Enzymes synthesizing the carbohydrate epitope of α-dystroglycan in the ER and Golgi apparatus, ordered by their reaction sequence, starting from phosphotrisaccharide (M3 core) formation.
| code | official name | reaction product added to the chain (or function played) | donor substrate |
|---|---|---|---|
| POMT1 | protein O-mannosyltransferase 1 | mannose (Man) | dolichol monophosphate mannose (Dol-P-Man)a |
| POMT2 | protein O-mannosyltransferase 2 | mannose (Man) | dolichol monophosphate mannose (Dol-P-Man)a |
| POMGnT2 | protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) | N-acetylglucosamine (GlcNAc) | UDP-GlcNAc |
| B3GALNT2 | beta-1,3-N-acetylgalactosaminyltransferase 2 | N-acetylgalactosamine (GalNAc) | UDP-GalNAc |
| POMK | protein O-mannose kinase | phosphate at C6 position of O-mannose | ATP |
| FKTN | fukutin | ribitol 5-phosphate (RboP) | CDP-ribitolb |
| POMGnT1 | protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) | binds to the growing saccharide chain and FKTN via its stem domain | — |
| FKRP | fukutin-related protein | ribitol 5-phosphate (RboP) | CDP-ribitolb |
| RXYLT1 | ribitol xylosyltransferase 1 | xylose (Xyl) | UDP-Xyl |
| B4GAT1 | beta-1,4-glucuronyltransferase 1 | glucuronic acid (GlcA) | UDP-GlcA |
| LARGE1 | LARGE xylosyl- and glucuronyltransferase 1 | [Xyl-GlcA]n | UDP-Xyl, UDP-GlcA |
| LARGE2 | LARGE xylosyl- and glucuronyltransferase 2 | [Xyl-GlcA]n | UDP-Xyl, UDP-GlcA |
| HNK-1ST | sulfotransferase or CHST10 carbohydrate sulfotransferase 10 | sulfate at C3 position of terminal GlcA | 3′-phosphoadenosine 5′-phosphosulfate (PAPS) |
aProvided in the cytosol by Dol-P-Man biosynthesis pathway including DPM1/2/3 (belonging to the dolichol-phosphate mannosyltransferase (DPM) Complex) and DOLK (dolichol kinase). Mutations in the gene encoding GDP-mannose pyrophosphorylase B (GMPPB), which functions in GDP-Man formation from Man-1-phosphate and GTP, were also reported in DGpathy patients [32].
bProvided in the cytosol by ISPD (isoprenoid synthase domain containing) that uses CTP and RboP (produced by an unknown enzyme) as substrates. ISPD is a cyitidyltransferase or CDP-ribitol pyrophosphorylase [33].
1.1. The M3 O-mannose core structure is formed in the endoplasmic reticulum at specific Thr residues (enzymes involved: POMT1/2, POMGnT2, B3GALNT2 and POMK)
The initial mannosylation reaction of α-DG is carried out by POMT, with POMT1 and POMT2 both needed for protein O-mannosyltransferase activity [34,35]. Site mapping studies have identified only two positions on α-DG for mannosylation by POMT1/2 to originate M3 core structures, namely Thr-317 and Thr-379, although some evidence suggests 319 and 381 may also be sites of M3 modification [17].
Mutations in these enzymes can cause autosomal recessive limb-girdle muscular dystrophies, LGMD2 K and LGMD2N [36], or severe Walker–Warburg syndrome (WWS), which is an autosomal recessive condition characterized by congenital muscular dystrophy, structural brain defects and eye malformations [34,35].
POMGnT2 (but not POMGnT1) is then responsible for starting the elongation of M3 in the ER [37], via the addition of β4 GlcNAc to the nascent sugar chain [17,27]. From a spatial-temporal perspective, the O-Man-modified α-DG precursor molecule encounters POMGnT2 in the ER first and POMGnT1 only later in the cis-Golgi, where its preferential activity leads to the formation of M1 O-mannose glycans [19] (see below). This points to POMGNT2 possessing specific substrate selectivity beyond simple recognition of an O-Man-modified amino acid, that univocally results in the formation of the M3 core structure [17]. Some of the mutations of POMGnT2 cause severe WWS [38], but others only result in mild forms of limb-girdle muscular dystrophy [39,40], while POMGnT2 knockout mice represent models for cobblestone lissencephaly [41].
The subsequent β3 addition of GalNAc to the growing chain is carried out in the ER by B3GALNT2 [42]. However, B3GALNT2 activity does not seem to fully correlate with the severity of the observed muscular dystrophy phenotypes [43]. The last step of this ER enzymatic phase is carried out by POMK (SGK196), a kinase that works with ATP as a donor substrate and is involved in the phosphorylation of the M3 glycan mannose on its C-6 position [44]. Three-dimensional structures of POMK have been solved [45,46], and it was recently shown that LARGE1 (see paragraph 1.3) can work efficiently only when POMK is active [22].
1.2. A single tandem of ribitol-phosphates is added in the Golgi (enzymes involved: FKTN, POMGnT1 and FKRP)
Fukutin (FKTN) catalyses the transfer of ribitol-phosphate (RboP) to α-DG using cytidine diphosphate ribitol (CDP-Rbo) as substrate. Among the enzymes of the pathway, FKTN is one of the most extensively characterized biophysically, despite the lack of high-resolution three-dimensional structural data [47–49]. Mutations in the FKTN gene can cause autosomal recessive limb-girdle muscular dystrophy (LGMD2M) [50], while a Dandy–Walker malformation was observed in the cerebellum [51]. The conditional knockout of FKTN in the mouse heart leads to a pathology that is observed only in later adulthood, when a severe cardiac dysfunction emerges [52]. It is worth to note that some missense mutations can instead lead to more severe consequences, such as a lethal case of WWS [53].
Interestingly and quite uniquely, POMGnT1 acts as a sort of ‘enzymatic chaperone’ in the fabrication of the M3 core structure. POMGnT1 binds FKTN via its stem domain and then facilitates the enzymatic action (i.e. addition of RboP) of FKTN on the developing M3 core [18,54,55]. POMGnT1 is localized in the cis-Golgi via interaction with the Golgi phosphoprotein-3, GOLPH3 [56], and if it were not thus tethered to the Golgi apparatus, it would probably interfere with the ER-localized enzymatic action of POMGnT2. As a consequence, α-DG would then display a wrong pattern of glycosylation as far as the location (i.e. at the level of specific Thr residues) of the resulting M3 and M1 core structures are concerned. In fact, POMGnT1 has been appropriately defined as a ‘cross-core enzyme’ since it is responsible for fabricating, together with the glycosyltransferases MGAT5B/GnT-IX (Vb), the M1 and M2 core structures [17]. Human α-DG has at least 25 O-mannosylation sites, the majority of which are populated by core M1 and M2 glycan structures via the action of POMGnT1 (M1) followed by MGAT5B/GnT-IX (Vb) (M2). POMGnT1 knockout in mice causes a severe form of muscle–eye–brain diseases [57] as described in certain patients [58], although some specific mutations can cause the less severe autosomal recessive limb-girdle muscular dystrophy, LGMD20 [59].
Like FKTN, FKTN-related protein (FKRP) uses CDP-Rbo as substrate (table 1) and transfers a second consecutive RboP to α-DG [27,28,60]. The synergic activity of these two enzymes produces the tandem RboP unit (RboP–RboP) required for the subsequent synthesis, mediated by RXYLT1, B4GAT1 and LARGE, of the laminin-binding glycoepitope on the O-mannosyl glycan. Mutations of FKRP can cause autosomal recessive limb-girdle muscular dystrophy LGMD2I [61,62].
1.3. Matriglycan, the polymer that is recognized by the α-DG binding partners (enzymes involved: RXYLT1, B4GAT1, LARGE1/2 and HNK-1ST)
The matriglycan represents the laminin-binding glycoepitope recognized by laminin and other binding partners [4,20]. Upon the addition of the RboP dimer and prior to the synthesis of the matriglycan polymer, a dimer of xylole and glucuronic acid is added by RXYLT1, a ribitol xylosyltransferase 1 (ribitol β1,4-Xylosyltransferase) [63] and B4GAT1 (β-1,4-glucuronyltransferase 1) formerly known as B3GNT1 [64,65]. Then, LARGE1 adds the repeating disaccharide unit [-3Xyl-α1,3GlcAβ1-]n [66] by employing its double capacity as a xylosyltransferase (domain 1) and a glucuronyltransferase (domain 2) [67]: the tandemly repeated polymer of these disaccharide units thus generated is called matriglycan, [20]. The N-terminal domain of α-DG binds LARGE and it could act as a chaperone for directing the enzymatic activity of LARGE towards its own mucin-like domain [68,69]. LARGE2 is a paralogue of LARGE1 found in Vertebrates [70]. The elongation process ends with the 3-O-sulfation of the last glucuronic acid within matriglycan catalysed by the HNK-1ST sulfotransferase [71] that transfers a sulfate group to the non-reducing end GlcA of matriglycan and prevents extension by LARGE1 [21].
Given the role played by α-DG for muscle stability and muscular dystrophies, we believe that it is important to study the evolutionary implications of this set of enzymes that is crucially responsible for producing the glycoepitope at the basis of α-DG function. In addition, we believe it is equalto trace back the origin of the enzymatic machinery that produces the M3 glycoepitope because the carbohydrate moieties of α-DG are hijacked by some viruses to enter eukaryotic cells [72,73].
2. Methods
2.1. Sequence comparison and bioinformatic methods
All the details referring to the human sequences used as ‘baits’ to find orthologous sequences of the 13 enzymes in the different animal lineages are reported in table 1. The ‘BLAST’ (Basic Local Alignment Research Tool) resource (specifically blastp, protein–protein BLAST), freely available at NCBI, has been used for all the searches, except that for Oscarella carmela belonging to the Homoscleromorpha sponges (Porifera), where the one available at Compagen (www.compagen.org) has been employed. Some additional sequence comparison searches have been performed at the web resource Wormbase (https://wormbase.org) for Caenhorabditis elegans. Electronic supplementary material is presented in a single file and includes all the sequence comparison details with the best scores found, enzyme by enzyme and group by group. The protein alignments have been carried out in the multiple sequence alignment resource muscle available at EMBL-EBI (https://www.ebi.ac.uk).
Given the genetic importance of the fruit fly, Drosophila melanogaster (Insecta, Diptera), for the study of muscle degeneration (see Results and discussion), some relevant orthologues are reported for Drosophilidae. Both for their genetic and evolutionary importance as well as for their wide use as model organisms, orthologues are also reported for the starlet sea anemone, Nematostella vectensis (Cnidaria, Anthozoa), for the Caenorhabditis genus which includes the widely studied worm C. elegans, for zebrafish, Danio rerio (Teleostei, Actinopterygii), and for the house mouse, Mus musculus (Mammalia, Rodentia).
3. Results and discussion
3.1. Homology of orthologous enzymes with their human counterparts
O-mannosylation, that was originally discovered in fungi [74,75], is known to be conserved from bacteria to humans [76] and multiple O-man glycosylation pathways have been found in eukaryotes [14]. O-mannosylation as such is not characteristic of plants [14], nevertheless very good hits with POMT1 and POMT2 have been found in Quercus suber (XP_023912063.1), Carpinus fangiana (KAB8356666.1) or Rhodamnia argentea (XP_030536296.1). The most likely hypothesis is that such hits originate from contamination with some pathogenic fungi when the plant sequence analysis was performed and deposited in the data bank [77] or, less probably, could represent cases of horizontal gene transfer (HGT) [78]. Indeed, the hit found in Quercus saber for example (XP_023912063, 784 aa) is practically identical to a glycosyltransferase from the fungus Baudoinia panamericana (XP_007679195, 791 aa, 83% identity on a 100% query cover), and many other very good matches are found with similar sequences belonging to other species of fungi, strongly suggesting a ‘cross-contamination’ of fungi sequences into plant sequences within the NCBI databank.
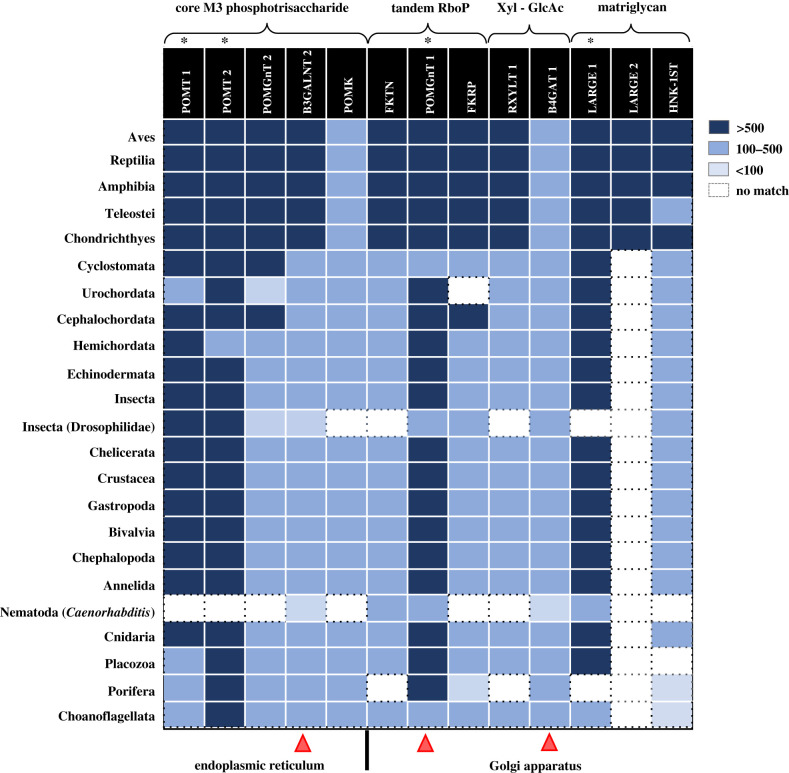
The sequences identified in free-living unicellular and colonial flagellate eukaryotic Choanoflagellata and the other metazoan lineages (all displaying with one of the best total scores) have been reported in the electronic supplementary material. All the enzymes (except LARGE2 that originates from a duplication first observed in Chondrichthyes; figure 2; see below) pre-date DG because they are all present in the nearest free-living single-celled eukaryotic precursor of Metazoa, Choanoflagellata where DG is not present [5]. Among basal Metazoa, in the ‘static’ Porifera, some of the 13 enzymes are not present (figure 2). An even higher degree of divergence and loss of significant matching sequences has been observed in the Caenorhabditis genus of Nematoda (in which only 5/13 orthologues are found). With the above exception, the enzymes presence is remarkably consistent starting from other basal metazoan, like in basal Placozoa where all the enzymes have been identified (except the sulfotransferase HNK-ST1 and LARGE2) (11/13). It is worth of note that HNK-ST1 was first found in another basal metazoan group such as Cnidaria (12/13). From Annelida upwards, all the enzymes are present except LARGE2. In Urochordata, POMGnT2 and FKRP ‘disappear’, and it remains unclear whether or not there is a paralogue of LARGE1 (figure 2; electronic supplementary material) as reported below in paragraph 3.3.
Figure 2.
Animal groups versus enzymes box showing the relevant similarities found between the human orthologous sequence baits for the thirteen enzymes (see table 2 for details) and the different animal lineages. The score reported (see electronic supplementary material for the exact values) is the ‘Max score’, i.e. the highest alignment score (bit-score) between the query sequence and the sequence segment found within the database. Score code: greater than 500, navy blue; 100–500, blue; less than 100, pale blue; and no significant match, white. For the matching details (including sequences codes and scores) between the human orthologues and the various groups, see electronic supplementary material. The asterisks mark the four enzymes that are more conserved (POMT1, POMT2, POMGnT1 and LARGE1) while the red triangles highlight B3GALNT2, POMGnT1 and B4GAT1, for which orthologues are present in all the lineages analysed.
3.2. Conservation and specificity of the M3 core enzymatic cascade
The most conserved enzymes with respect to the human ‘bait’ sequences used in this analysis (table 2) are POMT1/POMT2, POMGnT1 and LARGE1. It is perhaps not a coincidence that some of the more serious forms of dystroglycanopathy observed in human patients (displaying WWS or muscle–eye–brain disease) originate from mutations in some of these enzymes. This seems to be in line with what observed in mouse knockout model systems. For example, mutations in human POMGnT1 cause LGMD2O (late childhood, progression moderate) while mice are viable but affected by a severe form of muscle–eye–brain disease [57]. On the other hand, POMGnT2 knockout mice present with severe multiple phenotypes during embryonic development and perinatal death. [41,79].
Table 2.
Details of the human enzymes used as baits to identify orthologues in the different animal lineages by sequence comparison.
| name | location | chr. | length (a.a.) | code |
|---|---|---|---|---|
| POMT1a | ER | 9 | 747 | Q9Y6A1 |
| POMT2a | ER | 14 | 750 | Q9UKY4 |
| POMGnT2 | ER | 3 | 580 | Q8NAT1 |
| B3GALNT2 | ER | 1 | 500 | Q8NCR0 |
| POMK | ER | 8 | 350 | Q9H5K3 |
| FKTN | CGC/MGC | 9 | 461 | O75072 |
| POMGnT1a | CGC/MGC | 1 | 660 | Q8WZA1 |
| FKRP | CGC/MGC | 19 | 495 | Q9H9S5 |
| RXYLT1 | Golgi | 12 | 443 | Q9Y2B1 |
| B4GAT1 | Golgi | 11 | 415 | O43505 |
| LARGE1 | Golgi | 22 | 756 | O95461 |
| LARGE2 | Golgi | 11 | 721 | Q8N3Y3 |
| HNK-1STb | Golgi | 2 | 356 | O43529 |
ainvolved also in the formation of core M1 and M2.
binvolved also in the formation of core M1. chr.: chromosome location of the gene. CGC: cis-Golgi compartment, MGC: median-Golgi compartment.
Irrespective of the overall degree of conservation, our analysis shows how POMGnT1, B3GALNT2 and B4GAT1 are the only enzymes always present in all the groups considered (figure 2). It is important to remind that POMGnT1, along with POMT1 and many other known and still unidentified enzymes, is also involved in the formation of the abundant M1/M2 cores [17,19,44,80].
Have the enzymes responsible for the synthesis of the carbohydrate structure including M3, the two RboP, the additional Xyl-GlAc and sulfated Matriglycan evolved specifically for the glycosylation of α-DG? The most obvious answer to this question is no, as the full set of enzymes displaying a relevant degree of conservation to their orthologous human sequences is present in the Choanoflagellata (figure 2), which are single-cell eukaryotes precursors of multicellular organisms where DG cannot be found [5]. Therefore, a possible scenario is that the novel DG must have ‘hijacked’ an ancient pool of already available set of enzymes to fulfill its own ‘sugar decorating purposes'. A related question arises. Is the ‘M3 epitope’ shared by other proteins? In other words, do these enzymes have substrates other than α-DG? Although this remains to be established, α-DG is the only O-mannosylated protein that has been extensively studied so far [80]; therefore, the possible existence of other proteins containing very similar M3 core structures cannot be ruled out. Recently, it has been reported that indeed FKRP directs sialylation of fibronectin at the muscle basement membrane, influencing the overall stability of sarcolemma in muscular dystrophy and therefore strongly suggesting the existence of multiple target/pathways, alternative to DG, affected by the ‘M3’ enzymes [81]
The very high degree of conservation observed in Chordata strengthens the evidence that these 13 enzymes are important for muscle stability (figure 2). All considered, such remarkable conservation of the enzymatic cascade underlies the overall importance of α-DG carbohydrates for its functional activity. The emergence of the full pathway in early metazoan groups such as Placozoa and Cnidaria (well before the rise of more complex bilaterians) may suggest the crucial importance of a fully functional α-DG for stronger connections between the cytoskeleton and the muscular fibres that are needed for a general improvement in movement and locomotion, as already observed in Annelida and Mollusca (especially in Cephalopoda).
In Porifera, the overall degree of conservation is low. This raises the question of the possible existence of an ‘inverse correlation’ between the conservation of an enzymatic cascade that is fundamental for muscle stability and muscle-driven locomotion skills, and the staticity of Porifera in their adult stage. In fact, most sponges have a biphasic life cycle, with a planktonic phase (as moving larvae) and a benthic, less mobile one. Although it might sound like a fascinating hypothesis, it remains unclear if the high degree of divergence observed in sponges is directly related to their life cycle aspects. DG is already present in sponges, but both its degree of glycosylation and its molecular structure are currently scarcely characterized. Indeed, the presence of a region rich in Ser and Thr could have represented an ancestral mucin-like region in DG from Porifera, and mucin-like regions, although much shorter than in vertebrates, are already present in Placozoa and Cnidaria [5]. In Metazoa, laminins started to be present from Porifera and therefore represent binding partners for the α-DG carbohydrate scaffold, leading to the establishment of the high-affinity axis laminin (ECM)—DG sugar platform (membrane)—cytoskeleton that proved to be extremely successful from an evolutionary point of view. As a matter of fact, in the majority of metazoan groups (starting from Mollusca and Annelida), there is a consolidated presence of all the DG-binding partners at the cellular–matrix interface. Aside from detecting conserved orthologues of the relevant enzymes involved, a great deal of biochemical and structural work is granted to elucidate the exact carbohydrate moieties present in the orthologous DG complexes in the various invertebrate lineages [82].
Although the overall degree of glycosylation of α-DG in Porifera remains unclear, several enzymes of the glycosylation cascade are lost (4/13). As already stated, adult Porifera are static organisms and this has possibly favoured an hypoglycosylated α-DG, whereby the development of a specific multi-enzymatic cascade leading to the high level of α-DG glycosylation necessary to confer the muscle stability that movement requires was not needed. However, one necessary disclaimer is that a relatively low amount of sequence information is available for Porifera (i.e. there is still a limited variability of species sequenced) as well as for Placozoa, a group that includes only three genera (Trichoplax adhaerens, Hoilungia hongkongensis and Polyplacotoma mediterranea, with all the current sequence information available only for the former, see electronic supplementary material) and therefore caution is needed when drawing ultimate conclusions for these groups.
Important proteomic work from the Shcherbata's laboratory on Drosophila's DG corroborated and outlined the importance of the DG axis for muscle stability and neuronal development in the fruit fly, underlining its crucial role as a model system for the study of muscular dystrophies as well [83–86]. Still, it is very interesting to note that while in the class Insecta, the cascade appears to be well conserved, some relevant divergences seem to emerge in the family Drosophilidae (order Diptera). For example, while POMT1 and POMT2 have strong orthologues, whose mutations lead to the well-characterized twisted abdomen phenotypes [87,88], POMK, FKTN and RXYLT1 do not appear to have orthologues, and POMGnT2 does show only a limited similarity, with the EGF domain-specific O-linked N-acetylglucosamine transferase (a match with a score less than 100 was found for Drosophila grimshawi, XP_001989043). Moreover, also the widely and well-conserved B3GALNT2 and POMGnT1 are much less conserved in Drosophilidae as compared to other insect species. In addition, the same match found with B4GAT1/beta-1,4-glucuronyltransferase 1 was identified when using human LARGE1 and LARGE2 as bait (all the sequences details and alignment scores can be found in the electronic supplementary material). Together these observations suggest that there might be some differences within the M3 glycoepitope of Drosophila's DG relative to other Insecta [89], a fascinating hypothesis that awaits targeted experimental work in order to be verified.
3.3. Suppression of the M3 enzymatic cascade: a remarkable evolutionary divergence is apparent in the Caenorhabditis genus
Nematoda (roundworms) are known to encompass several parasite genera recognized as pathogens for plants and animals (including vertebrates and humans). Indeed, gene loss events frequently derive from a parasitic lifestyle. Interestingly, a DG orthologue is present in the well-characterized and non-parasitic nematode genus Caenorhabditis [90], although no extensive α-DG glycosylation shell has been found [5]. Strikingly, in this genus, most of the ‘M3 core’ enzymes are lost (only 5/13 identified; figure 2).
As far as the M3 enzymatic cascade is concerned, the Caenorhabditis genus seems to represent an extreme case of evolutionary divergence. This analysis revealed that most of the glycosylating enzymes do not have orthologues in the genus (only 5/13 are conserved, see below). Therefore, it seems unlikely that a ‘full and conventional’ M3 epitope is ever synthesized in those animals. Most relevantly, POMT1 and POMT2 have not been identified (like in plants) and therefore the required initial O-mannosylation step cannot take place.
Since the M3 enzymatic cascade is impaired, POMGnT1 and LARGE1 must be involved in other relevant glycosylation pathways. Consequently, it remains highly unclear what kind of carbohydrates (if any) would be present on the DG orthologous protein, also considering the evidence, emerged from our previous analysis, that the central ‘mucin-like’ highly glycosylated domain is missing in Caenorabditis [5,6]. As a matter of fact, the orthologue of DG we identified (i) does not share a mucin-like region and (ii) does not have, within its N-terminal region, the conserved S6 domain believed to be important for interacting with LARGE1 during the maturation of α-DG in the Golgi [68,69]. Interestingly, the knockout of the DAG1 orthologue in C.elegans gives rise to a phenotype in the vulvar epithelia as well as in the excretory cell epithelia and in motoneuron axon guidance, but does not seem to affect muscle or be dependent on dystrophin [91].
In members belonging to the Caenorhabditis genus, there are only orthologues for B3GALNT2, FKTN, POMGnT1, B4GALT1 and LARGE1. Based on the matches found using the human bait sequences (see details in the electronic supplementary material), it is possible to find the respective five orthologous genes of C. elegans (sqv-2, T07A5.1, M70.4a, bgnt-1.2 and lge-1). M70.4a is an orthologue of human FAM3A (FAM3 metabolism-regulating signalling molecule A) and FAM3C (FAM3 metabolism-regulating signalling molecule C) that harbour a domain that was recently shown to belong to POMGnT1 [55]. Sqv-2 and sqv-6 have been shown to correspond to human galactosyltransferases (sqv-2) and xylosyltransferases (sqv-6; although not similar to RXYLT1) [92].
It is interesting to note that these enzymes belong to a Golgi apparatus enzymatic machinery involved in the production of glycosaminoglycan chains (starting from xylose) and that sqv-2 and sqv-6 mutants display also a vulvar phenotype similar to that observed with the knockout of the DG-like gene of C. elegans that does not show a phenotype in muscle [91,92]. In Nematoda, there are no POMT1/POMT2 and O-mannosylation does not seem to be present; however, it is not known whether C. elegans DG could carry xylol-based carbohydrate chains instead. In general, these evidence point towards a slightly different role of the DG complex in Nematoda and to a different glycosylation pattern that would match the poorly conserved pattern of enzymes involved in the M3 core synthesis.
3.4. LARGE1 duplication in vertebrates
Contrary to what observed for other genes of the cascade that were already found duplicated ancestrally (POMT1/POMT2 in Choanoflagellata for example), the LARGE1 gene underwent a duplication event way more recently [70]. Genome duplication events contributed to increase the plasticity and complexity of vertebrate genomes, as observed especially in fish [93,94]. It was proposed that the redundancy in gene repertoires possessed by all vertebrates, including cyclostomes belonging to the agnathans (jawless fishes) lineage, was introduced primarily by two rounds of whole-genome duplications taking place during chordate evolution after the split of the Urochordata and Cephalochordata lineages, but before the radiation of gnathostomes (jawed vertebrates) [95].
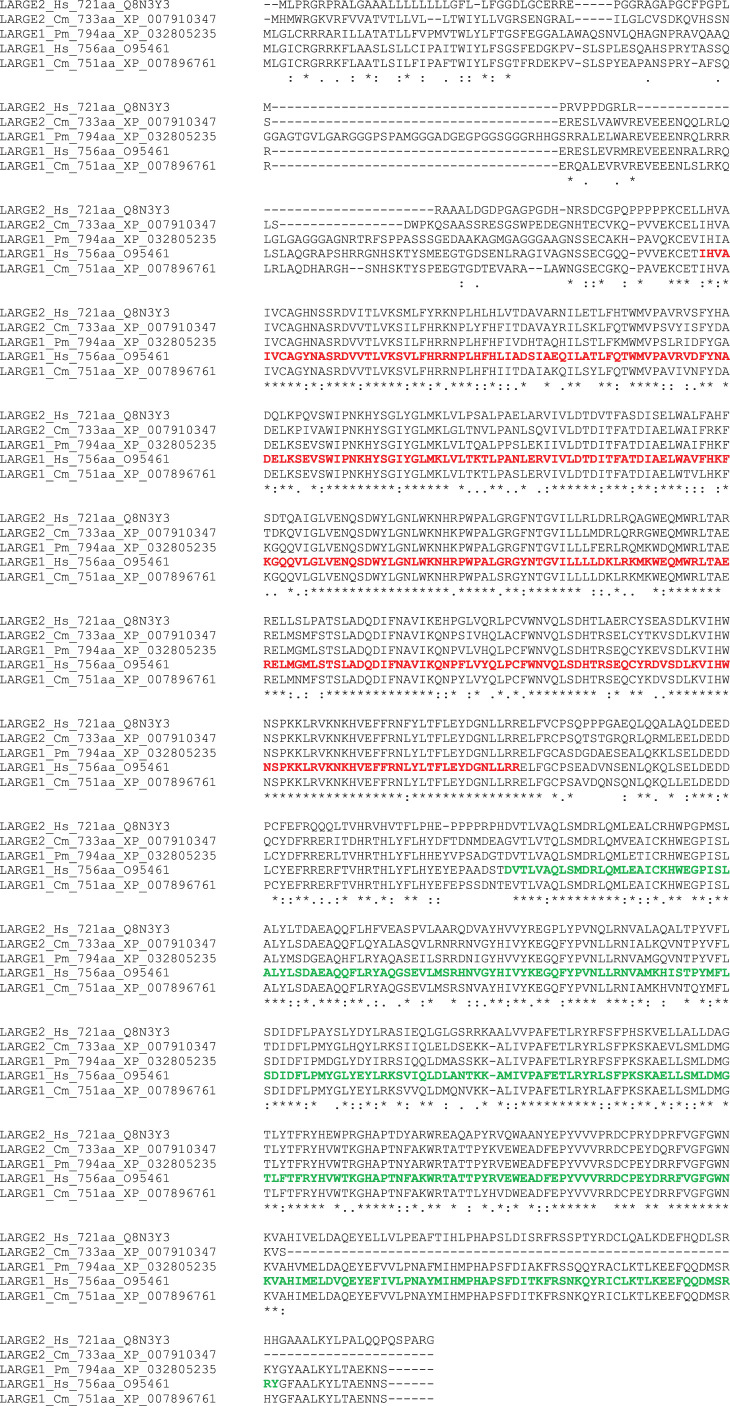
LARGE1 must be seen as one of the most crucial enzymes of the cascade since it is responsible for the synthesis of matriglycan, whose tandemly repeated disaccharide units are those specifically involved in the recognition by the so-called LG (laminin globular) domains harboured by DG-binding partners [4]. LARGE2 is a paralogue of LARGE1 with a demonstrated narrower tissue distribution in mice and with no significant expression in skeletal muscle [70], which shows the same enzymatic properties as LARGE1 [96]. The alignment between LARGE1 and LARGE2 from different species reported in figure 3 reveals a very high degree of homology, in which the conservation of the modules corresponding to the xylosyltransferase and the glucuronic acid transferase domains separated by a linker is confirmed [67].
Figure 3.
A multiple alignment of human LARGE1 and LARGE2 sequences with those from elephant shark (Chondrichthyes) and sea lamprey (Cyclostomata), showing the organization and conservation of the two subsequent catalytic domains oriented towards the Golgi lumen [70]. The two human LARGE1 catalytic domains are highlighted in red (xylosyltransferase domain) and green (glucuronyltransferase domain), respectively. Codes: Hs, Homo sapiens; Cm, Callorhinchus milii (elephant shark); Pm, Petromyzon marinus (lamprey). Asterisk: identical residues; colon: conserved substitutions; dot: semi-conserved substitutions.
It was previously observed that only the LARGE1 isoform was present in invertebrates and suggested that a gene duplication took place in vertebrates [70]. Indeed, this is in line with the results of the current analysis, which identified a paralogue of LARGE1, corresponding to LARGE2, only in vertebrates, namely in Chondrichthyes (although only observed in Callorhinchus milii, elephant shark, figure 3), Teleostei, Amphibia, Reptilia, Aves and Mammalia but not in Cyclostomata. It remains unclear whether LARGE1 was indeed duplicated in all Chondrichthyes, since LARGE2 could not be found in other species (as for example Carcharodon carcharias, white shark). Notably, the presence of additional significant matches in Ciona Intestinalis and Styela clava might suggest duplication also in Urochordata (see electronic supplementary material).
It was previously shown that two paralogues of DG can be found in the lamprey (Lethenteron japonicum) [5], and this is further confirmed by the two DG matches (XP_032827644, 939 a.a. and XP_032822703, 848 a.a.) found in Petrozon marinus at NCBI. We have observed also two paralogues of DG in species belonging to the Acanthomorpha lineage [97]. Therefore, these are likely to represent isolated events possibly independent from overall genome duplications [5,6].
The real physiological relevance of LARGE2 remains unclear, as knockout of LARGE2 in mice is phenotype-less [98], while knockout of LARGE1 generates a severe phenotype [99], in line with the high expression of LARGE1 in brain, heart and skeletal muscle [70]. However, evidence collected on LARGE2 point to its importance in the kidney [98], in prostate cancer [100,101], and in human colonic epithelium and colorectal cancer [102]. In addition, it was shown that LARGE2 can modify proteoglycans with the laminin-binding glycan [103]. All these results point towards a specific significance of LARGE2 rather than a mere functional redundancy. Therefore, in Cyclostomata, LARGE2 is likely to have undergone a selective inactivation, while in the other Chordata lineages the presence of a paralog gene is likely to have conferred an important degree of evolutionary advantage.
4. Conclusion
This initial reconstruction of the overall evolutionary pathway behind the rise of the M3 core structure and the DG/laminin axis implies that (i) all the enzymes of the M3 cascade have an ancestral origin that pre-dates DG and were already well consolidated before metazoan explosive rise; (ii) when DG came into play, at the level of basal Metazoa it was already able to hijack this pre-existing enzymatic machinery and prompt it to assemble into a specific DG-dedicated cascade starting with the initial O-mannosylation driven by POMT1/POMT2; (iii) the M3 cascade then enjoyed a prosperous evolutionary success in all the animal lineages, together with the DG/laminin axis it contributes to form. What was observed instead in Porifera and Nematoda is likely to represent examples of evolutionary divergence.
We believe that the high degree of conservation found in all the enzymes belonging to the M3 core structure cascade further stresses the importance of the enzymatic machinery responsible for DG sugar decoration and ultimately for the establishment of the DG-laminin axis, so fundamental to the achievement of a progressive muscle stability throughout all the Metazoan lineages. Such conservation might have conferred a significant evolutionary advantage, starting as early as from basal metazoans such as Placozoa and Cnidaria. This scenario fits with the idea of the evolutionary success of an integrated ‘molecular system’ of enzymes and their corresponding receptor-binding target system, able to accommodate new demanding biological, physiological and morphological requirements necessary for improving movement and locomotion skills (food collection, fighting, escaping from predators and so on).
Contributor Information
Maria Giulia Bigotti, Email: g.bigotti@bristol.ac.uk.
Andrea Brancaccio, Email: andrea.brancaccio@cnr.it.
Data accessibility
The data that support the findings of this study are all available within the manuscript and the electronic supplementary material [104].
Authors' contribution
A.B. conceived the project. M.G.B. and A.B. designed and executed the project and both wrote the paper. A.B. analysed the sequences, reviewed the literature and drafted the manuscript. M.G.B. contributed both to the preparation of the illustrations and to editing the final draft of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
M.G.B. is funded by the British Heart Foundation (grant no. CH/1/32804).
References
- 1.Sciandra F, Bigotti MG, Giardina B, Bozzi M, Brancaccio A. 2015. Genetic engineering of dystroglycan in animal models of muscular dystrophy. Biomed. Res. Int. 2015, 635792. ( 10.1155/2015/635792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickolls AR, Bönnemann CG. 2018. The roles of dystroglycan in the nervous system: insights from animal models of muscular dystrophy. Dis. Model. Mech. 11, dmm035931. ( 10.1242/dmm.035931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barresi R, Campbell KP. 2006. Dystroglycan: from biosynthesis to pathogenesis of human disease. J. Cell Sci. 119, 199-207. ( 10.1242/jcs.02814) [DOI] [PubMed] [Google Scholar]
- 4.Dempsey CE, Bigotti MG, Adams JC, Brancaccio A. 2019. Analysis of α-dystroglycan/LG domain binding modes: investigating protein motifs that regulate the affinity of isolated LG domains. Front. Mol. Biosci. 6, 18. ( 10.3389/fmolb.2019.00018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams JC, Brancaccio A. 2015. The evolution of the dystroglycan complex, a major mediator of muscle integrity. Biol. Open. 4, 1163-1179. ( 10.1242/bio.012468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brancaccio A, Adams JC. 2017. An evaluation of the evolution of the gene structure of dystroglycan. BMC Res. Notes 10, 19. ( 10.1186/s13104-016-2322-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancaccio A, Schulthess T, Gesemann M, Engel J. 1995. Electron microscopic evidence for a mucin-like region in chick muscle α-dystroglycan. FEBS Lett. 368, 139-142. ( 10.1016/0014-5793(95)00628-m) [DOI] [PubMed] [Google Scholar]
- 8.Bouchet-Séraphin C, Vuillaumier-Barrot S, Seta N. 2015. Dystroglycanopathies: about numerous genes involved in glycosylation of one single glycoprotein. J. Neuromuscul. Dis. 2, 27-38. ( 10.3233/JND-140047) [DOI] [PubMed] [Google Scholar]
- 9.Brancaccio A. 2019. A molecular overview of the primary dystroglycanopathies. J. Cell. Mol. Med. 23, 3058-3062. ( 10.1111/jcmm.14218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson RA, Henry MD, Daniels KJ, Hrstka RF, Lee JC, Sunada Y, Ibraghimov-Beskrovnaya O, Campbell KP. 1997. Dystroglycan is essential for early embryonic development: disruption of Reichert's membrane in Dag1-null mice. Hum. Mol. Genet. 6, 831-841. ( 10.1093/hmg/6.6.831) [DOI] [PubMed] [Google Scholar]
- 11.Riemersma M, et al. 2015. Absence of α- and β-dystroglycan is associated with Walker-Warburg syndrome. Neurology 84, 2177-2182. ( 10.1212/WNL.0000000000001615) [DOI] [PubMed] [Google Scholar]
- 12.Moremen KW, Tiemeyer M, Nairn AV. 2012. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448-462. ( 10.1038/nrm3383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. 1992. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696-702. ( 10.1038/355696a0) [DOI] [PubMed] [Google Scholar]
- 14.Larsen ISB, Narimatsu Y, Clausen H, Joshi HJ, Halim A. 2019. Multiple distinct O-mannosylation pathways in eukaryotes. Curr. Opin. Struct. Biol. 56, 171-178. ( 10.1016/j.sbi.2019.03.003) [DOI] [PubMed] [Google Scholar]
- 15.Manya H, Suzuki T, Akasaka-Manya K, Ishida HK, Mizuno M, Suzuki Y, Inazu T, Dohmae N, Endo T. 2007. Regulation of mammalian protein O-mannosylation: preferential amino acid sequence for O-mannose modification. J. Biol. Chem. 282, 20 200-20 206. ( 10.1074/jbc.M702369200) [DOI] [PubMed] [Google Scholar]
- 16.Hara Y, et al. 2011. Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc. Natl Acad. Sci. USA 108, 17 426-17 431. ( 10.1073/pnas.1114836108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halmo SM, Singh D, Patel S, Wang S, Edlin M, Boons GJ, Moremen KW, Live D, Wells L. 2017. Protein O-Linked Mannose β-1,4-N-Acetylglucosaminyl-transferase 2 (POMGNT2) is a gatekeeper enzyme for functional glycosylation of α-dystroglycan. J. Biol. Chem. 292, 2101-2109. ( 10.1074/jbc.M116.764712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuwabara N, et al. 2016. Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of α-dystroglycan. Proc. Natl Acad. Sci. USA 113, 9280-9285. ( 10.1073/pnas.1525545113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Meng C, Jin L, Chen X, Wang F, Cao H. 2015. Chemoenzymatic synthesis of α-dystroglycan core M1 O-mannose glycans. Chem. Commun. (Camb.) 51, 11 654-11 657. ( 10.1039/c5cc02913a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida-Moriguchi T, Campbell KP. 2015. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology 25, 702-713. ( 10.1093/glycob/cwv021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheikh MO, et al. 2020. HNK-1 sulfotransferase modulates α-dystroglycan glycosylation by 3-O-sulfation of glucuronic acid on matriglycan. Glycobiology 30, 817-829. ( 10.1093/glycob/cwaa024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walimbe AS, et al. 2020. POMK regulates dystroglycan function via LARGE1-mediated elongation of matriglycan. Elife 9, e61388. ( 10.7554/eLife.61388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciandra F, Bozzi M, Bigotti MG, Brancaccio A. 2013. The multiple affinities of α-dystroglycan. Curr. Protein Pept. Sci. 14, 626-634. ( 10.2174/1389203711209070644) [DOI] [PubMed] [Google Scholar]
- 24.Larson AA, et al. 2018. TRAPPC11 and GOSR2 mutations associate with hypoglycosylation of α-dystroglycan and muscular dystrophy. Skelet. Muscle 8, 17. ( 10.1186/s13395-018-0163-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu SH, Zhao P, Prabhakar PK, Sun T, Beedle A, Boons GJ, Moremen KW, Wells L, Steet R. 2018. Defective mucin-type glycosylation on α-dystroglycan in COG-deficient cells increases its susceptibility to bacterial proteases. J. Biol. Chem. 293, 14 534-14 544. ( 10.1074/jbc.RA118.003014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bozzi M, Di Stasio E, Scaglione GL, Desiderio C, Martelli C, Giardina B, Sciandra F, Brancaccio A.. 2013. Probing the stability of the ‘naked’ mucin-like domain of human α-dystroglycan. BMC Biochem. 14, 15. ( 10.1186/1471-2091-14-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Praissman JL, et al. 2016. The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife 5, e14473. ( 10.7554/eLife.14473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cataldi MP, Lu P, Blaeser A, Lu QL. 2018. Ribitol restores functionally glycosylated α-dystroglycan and improves muscle function in dystrophic FKRP-mutant mice. Nat. Commun. 9, 3448. ( 10.1038/s41467-018-05990-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imae R, et al. 2018. CDP-glycerol inhibits the synthesis of the functional O-mannosyl glycan of α-dystroglycan. J. Biol. Chem. 293, 12 186-12 198. ( 10.1074/jbc.RA118.003197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishihara R, Kobayashi K, Imae R, Tsumoto H, Manya H, Mizuno M, Kanagawa M, Endo T, Toda T. 2018. Cell endogenous activities of fukutin and FKRP coexist with the ribitol xylosyltransferase, TMEM5. Biochem. Biophys. Res. Commun. 497, 1025-1030. ( 10.1016/j.bbrc.2018.02.162) [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. 2019. A new patient-derived iPSC model for dystroglycanopathies validates a compound that increases glycosylation of α-dystroglycan. EMBO Rep. 20, e47967. ( 10.15252/embr.201947967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanagawa M, Toda T. 2018. Ribitol-phosphate-a newly identified posttranslational glycosylation unit in mammals: structure, modification enzymes and relationship to human diseases. J. Biochem. 163, 359-369. ( 10.1093/jb/mvy020) [DOI] [PubMed] [Google Scholar]
- 33.Riemersma M, et al. 2015. Human ISPD is a cytidyltransferase required for dystroglycan O-Mannosylation. Chem. Biol. 22, 1643-1652. ( 10.1016/j.chembiol.2015.10.014) [DOI] [PubMed] [Google Scholar]
- 34.van Reeuwijk J, et al. 2005. POMT2 mutations cause α-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J. Med. Genet. 42, 907-912. ( 10.1136/jmg.2005.031963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Reeuwijk J, et al. 2006. The expanding phenotype of POMT1 mutations: from Walker-Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum. Mutat. 27, 453-459. ( 10.1002/humu.20313) [DOI] [PubMed] [Google Scholar]
- 36.Nigro V, Savarese M. 2014. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 33, 1-12. [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa M, Nakamura N, Nakayama Y, Kurosaka A, Manya H, Kanagawa M, Endo T, Furukawa K, Okajima T. 2013. GTDC2 modifies O-mannosylated α-dystroglycan in the endoplasmic reticulum to generate N-acetyl glucosamine epitopes reactive with CTD110.6 antibody. Biochem. Biophys. Res. Commun. 440, 88-93. ( 10.1016/j.bbrc.2013.09.022) [DOI] [PubMed] [Google Scholar]
- 38.Manzini MC, et al. 2012. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am. J. Hum. Genet. 91, 541-547. ( 10.1016/j.ajhg.2012.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endo Y, et al. 2015. Milder forms of muscular dystrophy associated with POMGNT2 mutations. Neurol. Genet. 1, e33. ( 10.1212/NXG.0000000000000033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Endo T. 2019. Mammalian O-mannosyl glycans: biochemistry and glycopathology. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 95, 39-51. ( 10.2183/pjab.95.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa N, Yagi H, Kato K, Takematsu H, Oka S. 2015. Ectopic clustering of Cajal-Retzius and subplate cells is an initial pathological feature in Pomgnt2-knockout mice, a model of dystroglycanopathy. Sci. Rep. 5, 11163. ( 10.1038/srep11163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakane T, Angata K, Sato T, Kaji H, Narimatsu H. 2019. Identification of mammalian glycoproteins with type-I LacdiNAc structures synthesized by the glycosyltransferase B3GALNT2. J. Biol. Chem. 294, 7433-7444. ( 10.1074/jbc.RA118.006892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maroofian R, et al. 2017. B3GALNT2 mutations associated with non-syndromic autosomal recessive intellectual disability reveal a lack of genotype-phenotype associations in the muscular dystrophy-dystroglycanopathies. Genome Med. 9, 118. ( 10.1186/s13073-017-0505-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida-Moriguchi T, et al. 2013. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341, 896-899. ( 10.1126/science.1239951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Q, et al. 2016. Structure of protein O-mannose kinase reveals a unique active site architecture. Elife 5, e22238. ( 10.7554/eLife.22238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagae M, et al. 2017. 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy. Genes Cells 22, 348-359. ( 10.1111/gtc.12480) [DOI] [PubMed] [Google Scholar]
- 47.Holdbrook DA, Leung YM, Piggot TJ, Marius P, Williamson PT, Khalid S. 2010. Stability and membrane orientation of the fukutin transmembrane domain: a combined multiscale molecular dynamics and circular dichroism study. Biochemistry 49, 10 796-10 802. ( 10.1021/bi101743w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alhamidi M, Kjeldsen Buvang E, Fagerheim T, Brox V, Lindal S, Van Ghelue M, Nilssen Ø.. 2011. Fukutin-related protein resides in the Golgi cisternae of skeletal muscle fibres and forms disulfide-linked homodimers via an N-terminal interaction. PLoS ONE 6, e22968. ( 10.1371/journal.pone.0022968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marius P, Leung YM, Piggot TJ, Khalid S, Williamson PT. 2012. Probing the oligomeric state and interaction surfaces of Fukutin-I in dilauroylphosphatidylcholine bilayers. Eur. Biophys. J. 41, 199-207. ( 10.1007/s00249-011-0773-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachikawa M, Kanagawa M, Yu CC, Kobayashi K, Toda T. 2012. Mislocalization of fukutin protein by disease-causing missense mutations can be rescued with treatments directed at folding amelioration. J. Biol. Chem. 287, 8398-8406. ( 10.1074/jbc.M111.300905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traversa A, et al. 2020. Prenatal whole exome sequencing detects a new homozygous fukutin (FKTN) mutation in a fetus with an ultrasound suspicion of familial Dandy-Walker malformation. Mol. Genet. Genomic Med. 8, e1054. ( 10.1002/mgg3.1054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ujihara Y, Kanagawa M, Mohri S, Takatsu S, Kobayashi K, Toda T, Naruse K, Katanosaka Y. 2019. Elimination of fukutin reveals cellular and molecular pathomechanisms in muscular dystrophy-associated heart failure. Nat. Commun. 10, 5754. ( 10.1038/s41467-019-13623-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arora V, Bijarnia-Mahay S, Kulshreshtra S, Singh K, Puri RD, Verma IC. 2019. Prenatal presentation of a rare genetic disorder: a clinical, autopsy and molecular correlation. Autops. Case Rep. 9, e2019124. ( 10.4322/acr.2019.124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong H, et al. 2006. Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of alpha-dystroglycan. Biochem. Biophys. Res. Commun. 350, 935-941. ( 10.1016/j.bbrc.2006.09.129) [DOI] [PubMed] [Google Scholar]
- 55.Manya H, Kuwabara N, Kato R, Endo T. 2020. FAM3B/PANDER-like carbohydrate-binding domain in a glycosyltransferase, POMGnT1. Methods Mol. Biol. 2132, 609-619. ( 10.1007/978-1-0716-0430-4_52) [DOI] [PubMed] [Google Scholar]
- 56.Pereira NA, Pu HX, Goh H, Song Z. 2014. Golgi phosphoprotein 3 mediates the Golgi localization and function of protein O-linked mannose β-1,2-N-acetlyglucosaminyltransferase 1. J. Biol. Chem. 289, 14 762-14 770. ( 10.1074/jbc.M114.548305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Ball SL, Yang Y, Mei P, Zhang L, Shi H, Kaminski HJ, Lemmon VP, Hu H. 2006. A genetic model for muscle-eye-brain disease in mice lacking protein O-mannose 1,2-N-acetylglucosaminyltransferase (POMGnT1). Mech. Dev. 123, 228-240. ( 10.1016/j.mod.2005.12.003) [DOI] [PubMed] [Google Scholar]
- 58.Saredi S, et al. 2012. Novel POMGNT1 point mutations and intragenic rearrangements associated with muscle-eye-brain disease. J. Neurol. Sci. 318, 45-50. ( 10.1016/j.jns.2012.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raducu M, Baets J, Fano O, Van Coster R, Cruces J.. 2012. Promoter alteration causes transcriptional repression of the POMGNT1 gene in limb-girdle muscular dystrophy type 2O. Eur. J. Hum. Genet. 20, 945-952. ( 10.1038/ejhg.2012.40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuwabara N, et al. 2020. Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy. Nat. Commun. 11, 303. ( 10.1038/s41467-019-14220-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alhamidi M, Brox V, Stensland E, Liset M, Lindal S, Nilssen Ø. 2017. Limb girdle muscular dystrophy type 2I: no correlation between clinical severity, histopathology and glycosylated α-dystroglycan levels in patients homozygous for common FKRP mutation. Neuromuscul. Disord. 27, 619-626. ( 10.1016/j.nmd.2017.02.015) [DOI] [PubMed] [Google Scholar]
- 62.Henriques SF, Gicquel E, Marsolier J, Richard I. 2019. Functional and cellular localization diversity associated with Fukutin-related protein patient genetic variants. Hum. Mutat. 40, 1874-1885. ( 10.1002/humu.23827) [DOI] [PubMed] [Google Scholar]
- 63.Manya H, et al. 2016. The muscular dystrophy gene TMEM5 encodes a ribitol β1,4-xylosyltransferase required for the functional glycosylation of dystroglycan. J. Biol. Chem. 291, 24 618-24 627. ( 10.1074/jbc.M116.751917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer T, et al. 2014. The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated α-dystroglycan functional glycosylation. Elife 3, e03941. ( 10.7554/eLife.03941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Praissman JL, Live DH, Wang S, Ramiah A, Chinoy ZS, Boons GJ, Moremen KW, Wells L. 2014. B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of α-dystroglycan. Elife 3, e03943. ( 10.7554/eLife.03943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanagawa M, et al. 2004. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell 117, 953-964. ( 10.1016/j.cell.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 67.Righino B, Bozzi M, Pirolli D, Sciandra F, Bigotti MG, Brancaccio A, De Rosa MC.. 2020. Identification and modeling of a GT-A fold in the α-dystroglycan glycosylating enzyme LARGE1. J. Chem. Inf. Model. 60, 3145-3156. ( 10.1021/acs.jcim.0c00281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hara Y, et al. 2011. A dystroglycan mutation associated with limb-girdle muscular dystrophy. N. Engl. J. Med. 364, 939-946. ( 10.1056/NEJMoa1006939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bozzi M, Cassetta A, Covaceuszach S, Bigotti MG, Bannister S, Hübner W, Sciandra F, Lamba D, Brancaccio A. 2015. The structure of the T190M mutant of murine α-dystroglycan at high resolution: insight into the molecular basis of a primary dystroglycanopathy. PLoS ONE 10, e0124277. ( 10.1371/journal.pone.0124277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grewal PK, McLaughlan JM, Moore CJ, Browning CA, Hewitt JE. 2005. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology 15, 912-923. ( 10.1093/glycob/cwi094) [DOI] [PubMed] [Google Scholar]
- 71.Ong E, Yeh JC, Ding Y, Hindsgaul O, Fukuda M. 1998. Expression cloning of a human sulfotransferase that directs the synthesis of the HNK-1 glycan on the neural cell adhesion molecule and glycolipids. J. Biol. Chem. 273, 5190-5195. ( 10.1074/jbc.273.9.5190) [DOI] [PubMed] [Google Scholar]
- 72.Imperiali M, Thoma C, Pavoni E, Brancaccio A, Callewaert N, Oxenius A. 2005. O Mannosylation of α-dystroglycan is essential for lymphocytic choriomeningitis virus receptor function. J. Virol. 79, 14 297-14 308. ( 10.1128/JVI.79.22.14297-14308.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tayeh A, Tatard C, Kako-Ouraga S, Duplantier JM, Dobigny G. 2010. Rodent host cell/Lassa virus interactions: evolution and expression of α-Dystroglycan, LARGE-1 and LARGE-2 genes, with special emphasis on the Mastomys genus. Infect. Genet. Evol. 10, 1262-1270. ( 10.1016/j.meegid.2010.07.018) [DOI] [PubMed] [Google Scholar]
- 74.Strahl-Bolsinger S, Gentzsch M, Tanner W. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426, 297-307. ( 10.1016/s0304-4165(98)00131-7) [DOI] [PubMed] [Google Scholar]
- 75.Mouyna I, et al. 2010. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol. Microbiol. 76, 1205-1221. ( 10.1111/j.1365-2958.2010.07164.x) [DOI] [PubMed] [Google Scholar]
- 76.Lommel M, Strahl S. 2009. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19, 816-828. ( 10.1093/glycob/cwp066) [DOI] [PubMed] [Google Scholar]
- 77.Fernández-Alvarez A, Elías-Villalobos A, Ibeas JI. 2010. The requirement for protein O-mannosylation for Ustilago maydis virulence seems to be linked to intrinsic aspects of the infection process rather than an altered plant response. Plant Signal. Behav. 5, 412-414. ( 10.4161/psb.5.4.10805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, et al. 2020. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435. ( 10.1126/science.aba5435) [DOI] [PubMed] [Google Scholar]
- 79.Yagi H, et al. 2013. AGO61-dependent GlcNAc modification primes the formation of functional glycans on α-dystroglycan. Sci. Rep. 3, 3288. ( 10.1038/srep03288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng C, Sasmal A, Zhang Y, Gao T, Liu CC, Khan N, Varki A, Wang F, Cao H. 2018. Chemoenzymatic assembly of mammalian O-mannose glycans. Angew. Chem. Int. Ed. Engl. 57, 9003-9007. ( 10.1002/anie.201804373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood AJ, et al. 2021. FKRP-dependent glycosylation of fibronectin regulates muscle pathology in muscular dystrophy. Nat. Commun. 12, 2951. ( 10.1038/s41467-021-23217-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu F, Li D, Chen K. 2019. Structures and functions of invertebrate glycosylation. Open Biol. 9, 180 232. ( 10.1098/rsob.180232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kreipke RE, Kwon YV, Shcherbata HR, Ruohola-Baker H. 2017. Drosophila melanogaster as a model of muscle degeneration disorders. Curr. Top. Dev. Biol. 121, 83-109. ( 10.1016/bs.ctdb.2016.07.003) [DOI] [PubMed] [Google Scholar]
- 84.Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, Baker D, Ruohola-Baker H. 2007. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 26, 481-493. ( 10.1038/sj.emboj.7601503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yatsenko AS, Kucherenko MM, Xie Y, Aweida D, Urlaub H, Scheibe RJ, Cohen S, Shcherbata HR. 2020. Profiling of the muscle-specific dystroglycan interactome reveals the role of Hippo signaling in muscular dystrophy and age-dependent muscle atrophy. BMC Med. 18, 8. ( 10.1186/s12916-019-1478-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yatsenko AS, Kucherenko MM, Xie Y, Urlaub H, Shcherbata HR. 2021. Exocyst-mediated membrane trafficking of the lissencephaly-associated ECM receptor dystroglycan is required for proper brain compartmentalization. Elife 10, e63868. ( 10.7554/eLife.63868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ichimiya T, Manya H, Ohmae Y, Yoshida H, Takahashi K, Ueda R, Endo T, Nishihara S. 2004. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J. Biol. Chem. 279, 42 638-42 647. ( 10.1074/jbc.M404900200) [DOI] [PubMed] [Google Scholar]
- 88.Haines N, Seabrooke S, Stewart BA. 2007. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol. Biol. Cell 18, 4721-4730. ( 10.1091/mbc.e07-01-0047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schneider M, Baumgartner S. 2008. Differential expression of Dystroglycan-spliceforms with and without the mucin-like domain during Drosophila embryogenesis. Fly (Austin) 2, 29-35. ( 10.4161/fly.5726) [DOI] [PubMed] [Google Scholar]
- 90.Grisoni K, Martin E, Gieseler K, Mariol MC, Ségalat L. 2002. Genetic evidence for a dystrophin-glycoprotein complex (DGC) in Caenorhabditis elegans. Gene 294, 77-86. ( 10.1016/s0378-1119(02)00762-x) [DOI] [PubMed] [Google Scholar]
- 91.Johnson RP, Kang SH, Kramer JM. 2006. C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development 133, 1911-1921. ( 10.1242/dev.02363) [DOI] [PubMed] [Google Scholar]
- 92.Hwang HY, Olson SK, Brown JR, Esko JD, Horvitz HR. 2003. The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J. Biol. Chem. 278, 11 735-11 738. ( 10.1074/jbc.C200518200) [DOI] [PubMed] [Google Scholar]
- 93.Venkatesh B. 2003. Evolution and diversity of fish genomes. Curr. Opin. Genet. Dev. 13, 588-592. ( 10.1016/j.gde.2003.09.001) [DOI] [PubMed] [Google Scholar]
- 94.Volff JN. 2005. Genome evolution and biodiversity in teleost fish. Heredity (Edinb.) 94, 280-294. ( 10.1038/sj.hdy.6800635) [DOI] [PubMed] [Google Scholar]
- 95.Kuraku S, Meyer A, Kuratani S. 2009. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol. Biol. Evol. 26, 47-59. ( 10.1093/molbev/msn222) [DOI] [PubMed] [Google Scholar]
- 96.Inamori K, Hara Y, Willer T, Anderson ME, Zhu Z, Yoshida-Moriguchi T, Campbell KP. 2013. Xylosyl- and glucuronyltransferase functions of LARGE in α-dystroglycan modification are conserved in LARGE2. Glycobiology 23, 295-302. ( 10.1093/glycob/cws152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pavoni E, Cacchiarelli D, Tittarelli R, Orsini M, Galtieri A, Giardina B, Brancaccio A. 2007. Duplication of the dystroglycan gene in most branches of teleost fish. BMC Mol. Biol. 8, 34. ( 10.1186/1471-2199-8-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Inamori K, et al. 2014. Endogenous glucuronyltransferase activity of LARGE or LARGE2 required for functional modification of α-dystroglycan in cells and tissues. J. Biol. Chem. 289, 28 138-28 148. ( 10.1074/jbc.M114.597831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grewal PK, Holzfeind PJ, Bittner RE, Hewitt JE. 2001. Mutant glycosyltransferase and altered glycosylation of alpha-dystroglycan in the myodystrophy mouse. Nat. Genet. 28, 151-154. ( 10.1038/88865) [DOI] [PubMed] [Google Scholar]
- 100.Esser AK, et al. 2013. Loss of LARGE2 disrupts functional glycosylation of α-dystroglycan in prostate cancer. J. Biol. Chem. 288, 2132-2142. ( 10.1074/jbc.M112.432807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang Q, Miller MR, Schappet J, Henry MD. 2015. The glycosyltransferase LARGE2 is repressed by Snail and ZEB1 in prostate cancer. Cancer Biol. Ther. 16, 125-136. ( 10.4161/15384047.2014.987078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dietinger V, García de Durango CR, Wiechmann S, Boos SL, Michl M, Neumann J, Hermeking H, Kuster B, Jung P.. 2020. Wnt-driven LARGE2 mediates laminin-adhesive O-glycosylation in human colonic epithelial cells and colorectal cancer. Cell. Commun. Signal. 18, 102. ( 10.1186/s12964-020-00561-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Inamori KI, Beedle AM, de Bernabé DB, Wright ME, Campbell KP.. 2016. LARGE2-dependent glycosylation confers laminin-binding ability on proteoglycans. Glycobiology 26, 1284-1296. ( 10.1093/glycob/cww075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bigotti MG, Brancaccio A. 2021. High degree of conservation of the enzymes synthesizing the laminin-binding glycoepitope of α-dystroglycan. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bigotti MG, Brancaccio A. 2021. High degree of conservation of the enzymes synthesizing the laminin-binding glycoepitope of α-dystroglycan. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The data that support the findings of this study are all available within the manuscript and the electronic supplementary material [104].