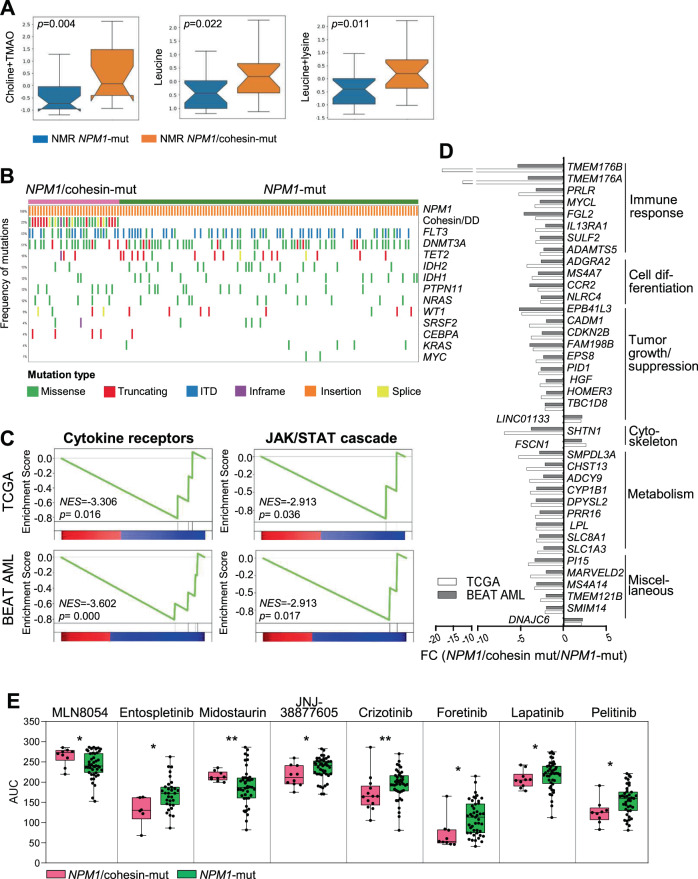
Fig. 6. Metabolic, genomic, transcriptomic and drug response differences between NPM1/cohesin-mut and NPM1-mut AML.
A Serum metabolites separating NPM1/cohesin-mut and NPM1-mut AML (TMAO trimethylamine-N-oxide). B Oncoprint of mutations in AML-related genes (frequency >3% in the overall population) in NPM1/cohesin-mut and NPM1-mut AML. WES data were obtained from the TCGA (n = 13 NPM1/cohesin-mut, n = 33 NPM1-mut) and BEAT AML (n = 19 NPM1/cohesin-mut, n = 72 NPM1-mut, including 7 relapse cases) cohorts. Rows denote genes or groups of genes (cohesin/DD cohesion/DNA damage-related genes). Columns represent frequency of mutations and single patients (ITD internal tandem duplication). C Signatures of cytokine receptors and JAK-STAT cascade from GSEA showing significance in both datasets (TCGA, left to right: cytokine–cytokine receptor binding, regulation of JAK-STAT cascade, n = 9 NPM1/cohesin-mut, n = 25 NPM1-mut; BEAT AML, left to right: cytokine receptor activity, JAK/STAT cascade, n = 14 NPM1/cohesin-mut, n = 47 NPM1-mut, including 3 relapse cases). D Genes involved in immune response, cell differentiation, tumor growth regulation, cytoskeleton, metabolism and other cellular processes, showing a significantly different expression between NPM1/cohesin-mut and NPM1-mut AML in both cohorts. E Area under the curve (AUC) for the drugs showing a significantly different response between NPM1/cohesin-mut and NPM1-mut AML was plotted for the two cohorts (NPM1/cohesin-mut, n = 6–13; NPM1-mut, n = 31–45) [1]: MLN8054 (Aurora kinase A inhibitor), Entospletinib (SYK inhibitor), Midostaurin (FLT3, JAK inhibitor), JNJ-38877605 (MET inhibitor), Crizotinib (ALK, MET, ROS1, NTRK inhibitor), Foretinib (MET, KDR, TIE inhibitor), Lapatinib (ErbB-2, EGFR inhibitor), Pelitinib (EGFR inhibitor). Boxes represent the mean (horizontal line) and extend from the 25th to 75th percentiles; whiskers extend from the minimum to the maximum value and each value is plotted (*p ≤ 0.05, **p ≤ 0.01).