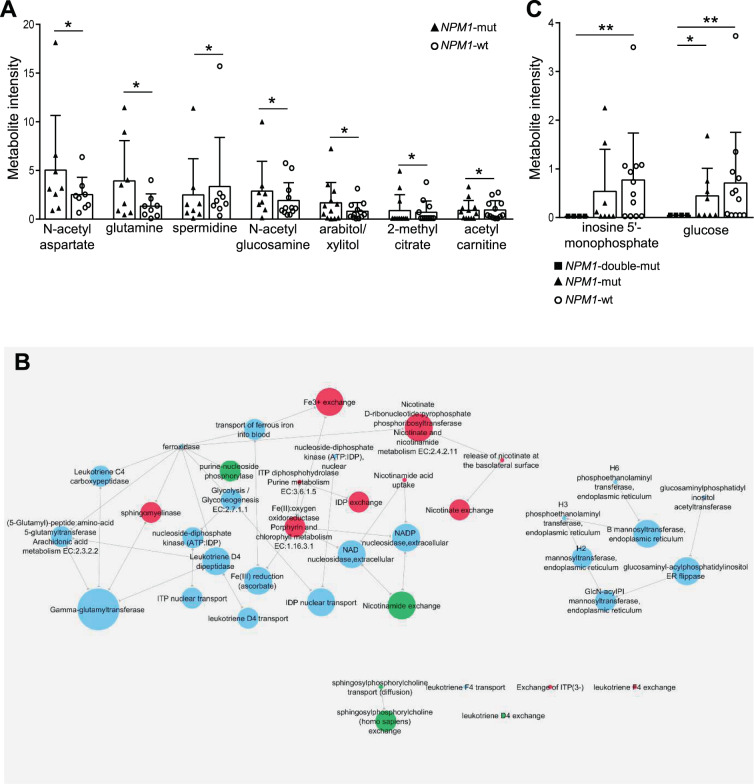
Fig. 7. Modeling the metabolic network of NPM1/cohesin-mut AML.
A Intracellular metabolites from FVA analysis showing a significant different intensity between NPM1-mut (n = 8) and NPM1-wt (n = 12, normal karyotype) in MS analysis. Metabolite intensity normalized on DNA concentration is shown in the plot. Significance was obtained by Welch t-test (*p ≤ 0.05). B NPM1/cohesin-mut specific metabolic reaction perturbation network. (red: minimum flux, green: maximum flux, light blue: no information among NPM1/cohesin-mut-specific alterations). Sizes of nodes are proportional to links originating from that node and pointing towards others (outdegree). C Intracellular metabolite intensity of NPM1/cohesin-mut-specific perturbations predicted by FVA (NPM1/cohesin-mut, n = 4; NPM1-mut, n = 8, NPM1-wt, n = 12, normal karyotype). Metabolite intensity normalized on DNA concentration is shown in the plot. Significance was obtained by Welch t-test (*p ≤ 0.05, **p ≤ 0.01).