Introduction
Interest in machine learning (ML) approaches to analyze patient data is in an explosive phase of growth. Underpinning this burgeoning interest are several advances in technology, including availability of computing power, evolution of ML software, and the ubiquity of electronic health records (EHRs). Personal computers now have sufficient computational power to run some ML algorithms for small to medium datasets, encompassing many of the datasets of interest in clinical medicine. Even for very large datasets (“big data”, with number of data points in the billions, trillions, or more), access to parallel computing resources is now widespread at academic medical centers and via cloud computing platforms. Advances in ML software maturity have substantially lowered the expertise threshold for ML. Many applications now exist, several of them free and open-source, assisting people at all skill levels to analyze their data using ML algorithms; code libraries to run ML algorithms are also available for use in Python,1 R,2 SAS,3 Stata,4 MATLAB5, and other programming languages. Importantly, most healthcare data are now stored digitally, within EHRs and other digital databases; in theory, these data may therefore be more easily collated and fed to ML algorithms on a large scale. The increasing adoption of digital technologies to collect patient data, such as wearables and online questionnaires, will only increase the appetite for ML implementation.6 These advances have led to a growing number of ML applications across a number of fields, yielding insights that would have previously been unobtainable or very difficult to obtain using “traditional” statistical methods.7,8 Analyses of healthcare datasets using ML are emerging with increasing frequency in the literature; within the realm of cancer research, ML approaches are driving deeper understanding of cancer risk, diagnosis, prognosis, and treatment (Table 1).
Table 1.
Example Studies Using Machine Learning in Cancer Research.
| Topic | Example Study | How did ML advance understanding? | Additional examples |
|---|---|---|---|
| Cancer risk | Comparison of machine learning risk model versus a validated clinical risk tool (BOADICEA) to calculate breast cancer risk using data from 112,587 patients27 |
|
Yala et al 2019,28 Nartowt et al 2019,29 Perera et al 202030 |
| Diagnosis | Development of a DNA methylation-based classification model for central nervous system tumor diagnosis, using 2,801 reference samples31 |
|
Wang et al 2019,23 Yamada et al 2019,32 Tschandl et al 201933 |
| Prognosis | Development of prognostic classification models for 5- year overall survival after surgery for pancreas cancer, using tissue samples from 194 patients34 |
|
Chen et al 2007,35 Wang et al 2011,36 Jiang et al 2018,37 Huang et al 201938 |
| Treatment | Development of predictive models for response to chemotherapy (gemcitabine [gem] and 5-fluorouracil [5- FU]) based on gene expression data from patients’ primary tumor tissue (n=92 patients, expression levels for 60,483 genes)39 |
|
Mucaki et al 2019,40 Cain et al 2018,41 Huang et al 201742 |
ML is generally used for classification or clustering of items that group together based on features (or variables) in large datasets. ML models can be utilized to identify patterns in data, or to build predictive models to be used for real-time outcome prediction or decision support. Advantages of ML include the ability to create models that mix multiple variable classes (i.e., continuous, categorical, and binary variables can be used within the same model), enhanced techniques for imputation of missing data, and in many cases, higher predictive precision compared to traditional predictive models. However, there is a well-described tradeoff in ML between predictive power (flexibility), and explanatory power (interpretability), both of which are crucial considerations for clinical questions. ML models with the highest predictive power are generally opaque with respect to identifying causal associations and explanatory mechanisms. High predictive power is desirable in the oncology clinic for accurate forecasting of clinical outcomes (e.g. survival, treatment benefit, tolerability, quality of life) and reducing patient uncertainty; however, interpretability may be necessary for pinpointing factors amenable to intervention and enhancing clinicians‟ acceptance and uptake of the model.
ML methods may be particularly suitable for analyzing data from older adults with cancer. By 2030, 70% of all cancer diagnoses will occur in people >65 years of age,9 but this same group comprises only 40% of participants in clinical trials which establish the standards for cancer treatment.10 Typically, older participants in clinical trials are the most “fit” representatives of their age groups, and therefore using the randomized clinical trial paradigm to guide therapy decisions will not sufficiently represent older adults with competing risks. Older adults have many potentially competing risks for adverse outcomes, creating difficulties in analyzing how treatments affect outcomes like overall survival within usual clinical trial models.11 Moreover, identifying the numerous variables impacting outcomes in older adults with cancer (including patient-reported outcomes, or PROs) has been the focus of extensive prior work in geriatric oncology,12 but these variables are often not captured or analyzed for clinical trials.
Pragmatic datasets such as EHR-abstracted data merged with patient-reported outcomes (PROs), and analyzed using novel ML methods, may overcome these knowledge limitations, including older, frailer adults not suitable for clinical trials, and broadening the set of available variables. As mentioned above, ML methods excel at mixing of categorical variables often found in PROs with continuous variables found in EHRs (e.g. weight, white blood cell count, hemoglobin) where traditional statistical methods may struggle. Importantly, in assembling these datasets, particularly those with high dimensionality (a large number of variables), data cleaning, model analysis, and model interpretation are critical and must be accomplished with care and expertise so as not to overstate conclusions. Successful blending of ML techniques with large pragmatic datasets may complement, and perhaps even overtake, clinical trials as a source of knowledge in this population. Additionally, these methods could permit better characterization and understanding of patient trajectories,13 leading to better tools to help with shared decision-making between oncologists and patients.
This primer introduces a series of methods articles about the use of ML approaches in geriatric oncology datasets. The purpose of these articles is to explain the steps in application of ML, discuss differences with traditional statistical approaches, and to compare model outputs in terms of flexibility, accuracy, and interpretability. Two subsequent papers will examine the use of ML to predict falls in older adults on chemotherapy, and to identify groupings of PROs that may co-occur and/or have predictive importance in this population.
Machine Learning: A brief primer
Machine learning algorithms are generally categorized as either supervised learning (usually known as „classification‟) or unsupervised learning (of which a common application is “clustering”). Supervised learning algorithms, such as k-nearest neighbor, random forest, and artificial neural networks, are trained on datasets with known, labeled outcomes of interest (e.g. frailty, survival). Supervised ML algorithms infer decision rules about how to assign or predict the class labels of new data points; for this reason, regression is often considered to be a supervised learning method, as it yields a model which can be used to classify or predict output from new sets of data points. Availability of validated (i.e. ground truth) labelled data is a requirement for supervised learning methods to succeed. For example, we trained supervised learning models to classify older adults who fell within 3 months of starting chemotherapy versus non-fallers (i.e., “faller” versus “non-faller” was the class label), using geriatric assessment, demographic, cancer-related, and other variables as model input. Description of the approach and model output will be the focus of the next paper in this methods series.
Unsupervised learning models explore unlabeled data to uncover the underlying data structure. It is most often used to detect previously unknown patterns within datasets. Most unsupervised ML algorithms are related to „clustering‟, which attempts to define clusters of data points which are “closer” or more similar to each other. This is easily accomplished and visualized in two or three dimensions (Figure 1), but given a high number of variables (and therefore dimensions), more sophisticated and computationally intensive algorithms14 and visualization methods15 are required. Unsupervised methods are often used in an exploratory way, rather than to yield definitive conclusions, and output is highly dependent on the algorithm and hyperparameters selected. Hyperparameters, also called tuning parameters, are values used to control the behavior of the ML algorithm (e.g., number of clusters, distance or density thresholds, type of linkage between clusters). Algorithms exist to detect clusters based on spatial distance between data points, space or subspace density, network connectivity between data points, or other measures; each of these techniques may “detect” markedly different cluster groupings. Biological taxonomies, such as species taxonomies, were developed using early conceptual precursors to unsupervised ML, and taxonomy problems today are often tackled using unsupervised ML. Within the medical field, clustering has been used in drug discovery,16 genomic analysis,17 and analysis of medical images,18 among other applications. The application of unsupervised ML to patient-reported data is in its infancy, but we have applied unsupervised ML algorithms to patient-reported symptom data to try to detect patterns within a dataset of older adults with advanced stage cancer receiving chemotherapy.19 This example will be the focus of a subsequent paper in this methods series.
Figure 1.
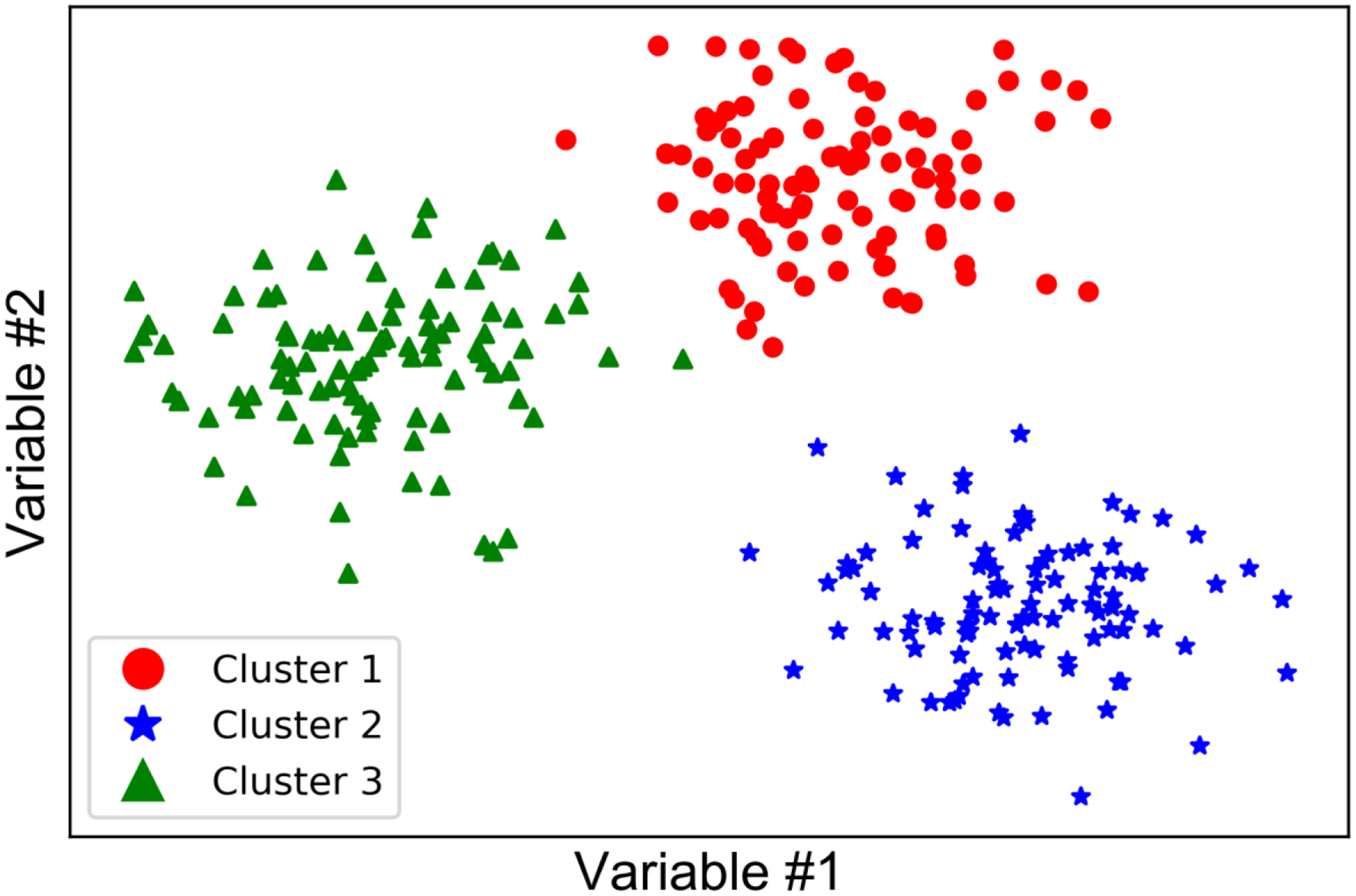
Visualizing clusters in 2 dimensions.
Supervised and unsupervised ML can be used effectively in tandem or in sequence depending on the particular dataset and research question. For example, unsupervised ML could be used to detect latent classes of patients, and then supervised learning models could be trained using these latent classes as the class labels. “Semi-supervised” approaches use a combination of the two techniques; for example, if labelling data points is resource-intensive, a few could be labelled while most remain unlabeled, and a semi-supervised approach would be valid. Other techniques under the rubric of ML include time series forecasting, with particular utilization by businesses and industries that analyze financial and sales data. Most healthcare data, including data obtained from patients with cancer, have a temporal component, and an understanding of trajectory is often a crucial component of cancer treatment decision-making. This may be particularly true for older adults, who often value knowledge about their anticipated functional, cognitive, or symptomatic trajectories above knowledge about survival outcomes.20
Machine Learning for Geriatric Oncology: Drawbacks and Challenges
Despite the promise entailed by ML methods for analyzing highly multivariable and/or longitudinal data collected from older adults with cancer, several limitations should temper enthusiasm and prompt careful scrutiny of these methods. For the foreseeable future, the lack of EHR interoperability and collaborative, shared data architecture may limit the assembly of datasets sufficiently large for ML analysis. Similar to traditional statistical methods like linear regression (a ML model itself), a sufficient sample size is necessary for input to ML algorithms, to minimize the competing risks of bias (causing underfitting of the data) and variance (causing overfitting). ML models can be “data hungry”: although heuristics to estimate necessary sample size are controversial, it is estimated that some common ML algorithms may require more than 10 times the number of samples per variable compared to linear regression models.21 Even the largest datasets typically collected in prospective clinical trials, with sample sizes in the low hundreds, may be insufficient for ML analysis; other large databases like SEER-Medicare do not collect all data relevant to older adults (such as PROs and geriatric assessment data). Large prospective datasets in geriatric oncology are rare. In order to address this problem, multicenter prospective data collection of the relevant variables is needed. Attempts to define a standardized “minimum data set” for older adults with cancer, such as recent American Society of Clinical Oncology (ASCO) guidelines, can facilitate the development of large datasets.22 These data should optimally be maintained in a centralized, collaboratively resourced data warehouse with standardized intake and validation procedures.
Even with large datasets, and despite the availability of user-friendly software packages allowing anyone to apply ML methods, significant expertise is required to prepare the data for analysis, select and tune the appropriate ML algorithm, implement the model efficiently within the available computational infrastructure, and assess for potential bias. Data validation, data transformation (such as selecting variable type and normalizing variable values), and feature selection (choosing the variables to include in the model) require considerable time and experience. A variety of ML algorithms are available, and can yield significantly different output with the same training data set. Even within algorithms, model tuning (i.e., selecting hyperparameters) is not standardized and is based largely on expertise and experience. Unlike with regression methods, wherein the statistical model can be examined variable by variable, many ML algorithms are “black boxes,” with inscrutable behavior generating the model output. Even when the inner workings of the model can be inspected, as in regression, or with recently developed mechanisms to examine the inner workings of neural network models,23,24 it can be very difficult even for experts in these methods to identify, describe, and correct for sources of bias. Ethically, it is crucial that research teams have the skill and dedication to rigorously examine, validate, and question the reliability and completeness of data input into ML models, particularly when the workings of the model cannot be easily parsed. Failure to eliminate sources of bias in the model training data can result in meaningless (at best) or highly destructive (at worst) conclusions.25
The role of “data scientist” is a relatively new concept, combining the totality of skills needed to work with data across its life-cycle (Figure 2). Among these skills are knowledge of data architecture (such as databases), programming ability, statistical knowledge (including deep understanding of ML algorithms), proficiency with data visualization, and content expertise. Unfortunately, although approximately 30% of stored data in the world is healthcare data, only 3% of data scientists in the United States work within the healthcare sector.26 It is estimated, by benchmarking with other data-driven industries, that 10 to 20 times more data scientists are needed in healthcare.26 The demand for these professionals far outpaces their availability, but their skills could greatly augment the analysis of complex data in older adults with cancer. Research teams in geriatric oncology should consider the addition of data scientists to their teams; at the University of Rochester, rich collaborations have formed between the Geriatric Oncology Research Group and the Goergen Institute for Data Science, yielding work on ML methods applied to geriatric oncology datasets.
Figure 2.
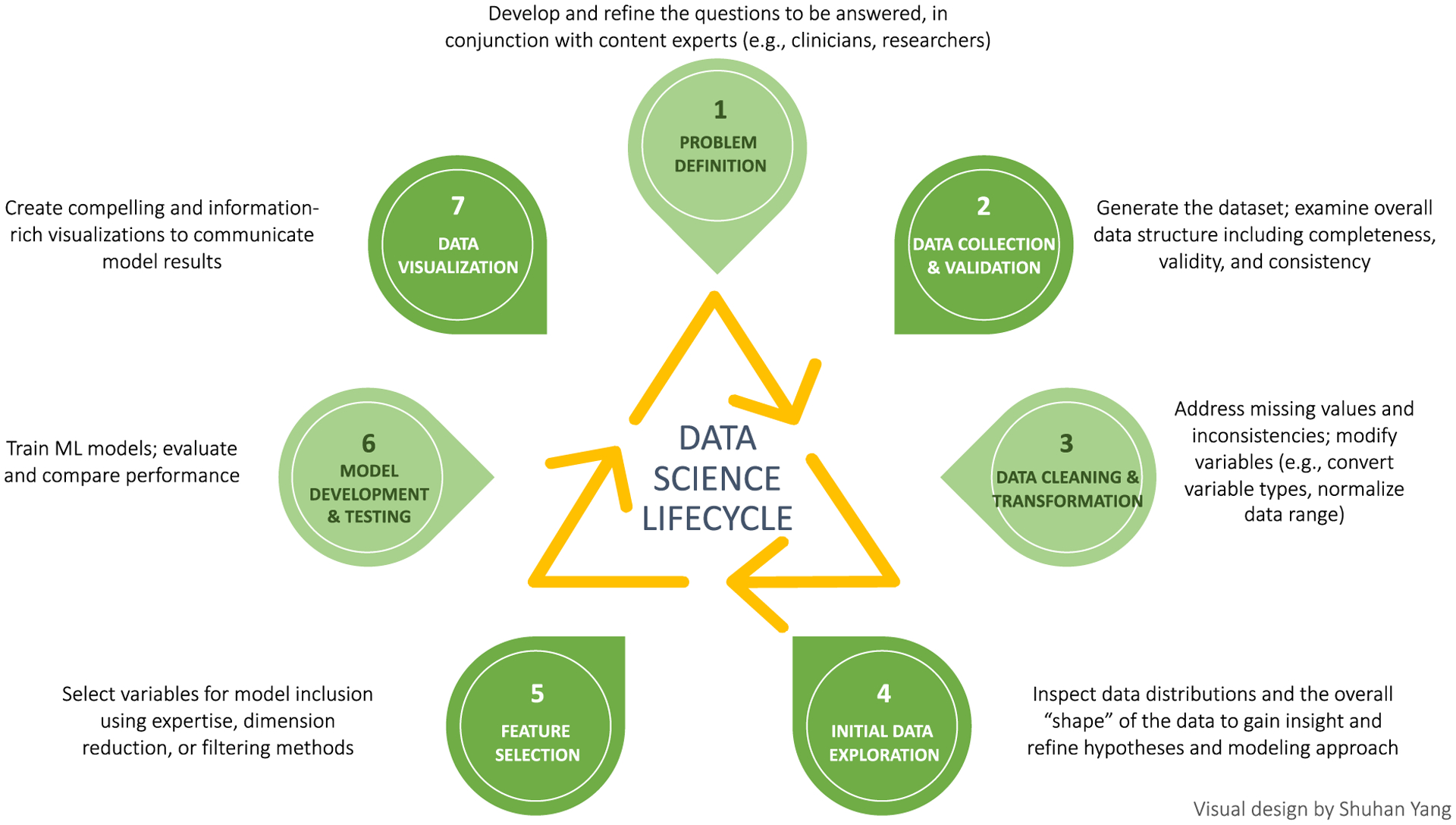
Data Science lifecycle.
Conclusion
Use of ML algorithms in the analysis of healthcare data is increasingly common, and clinicians should be familiar with basic ML concepts. Application of ML to understand trajectory and outcomes in older adults with cancer is intriguing, albeit with several constraints including small currently available clinical datasets and limited workforce with ML expertise within healthcare. These constraints have limited adoption of both “traditional” ML algorithms (the focus of this paper) as well as more sophisticated methods such as deep learning which are being pioneered in other industries. Deep learning methods can enhance model performance, but require massive datasets and computational power which are out of reach for most clinical applications at the current time (Figure 3).
Figure 3.
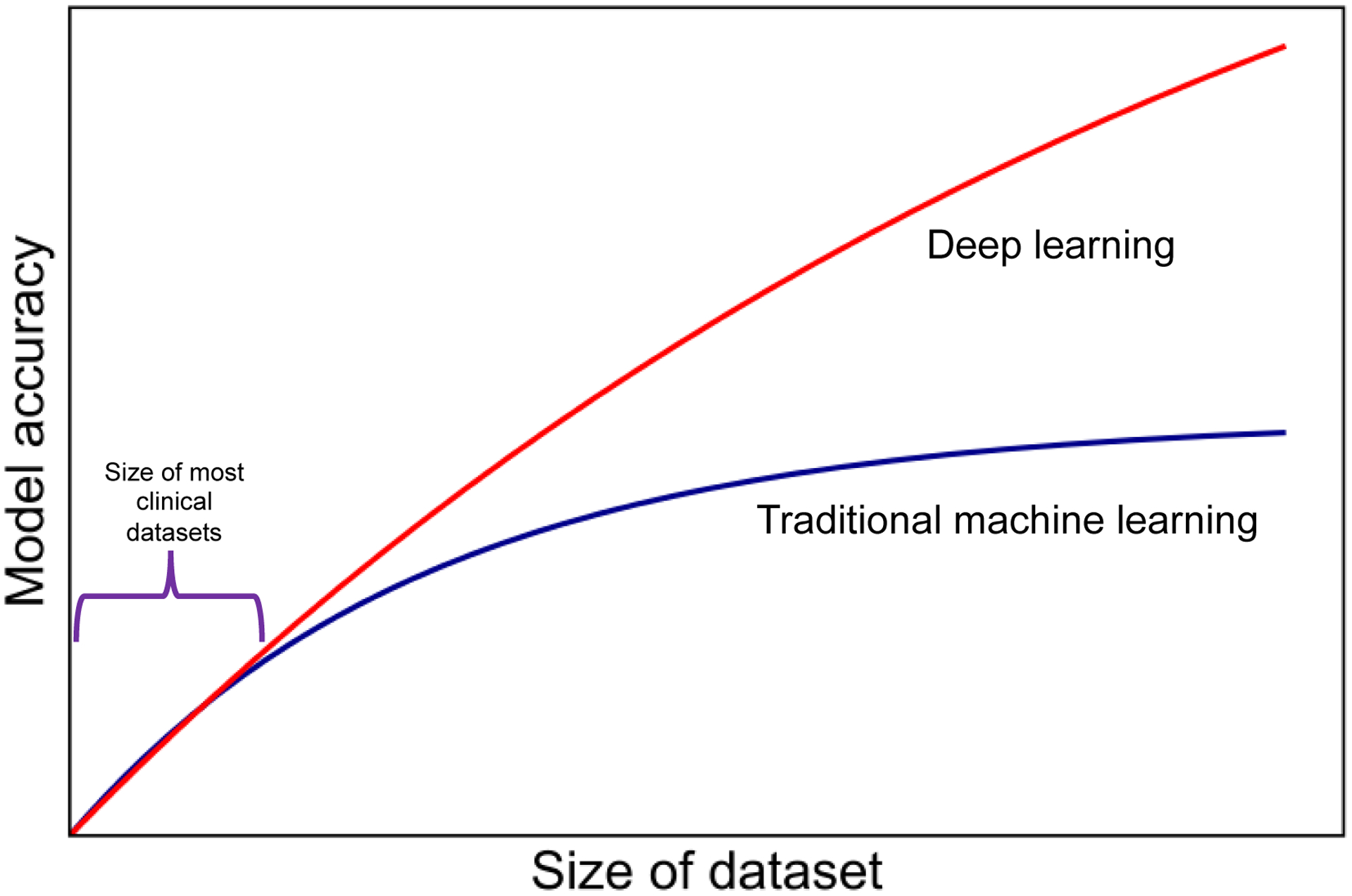
Model performance as a function of amount of data for traditional ML and deep learning algorithms.
This introduction to basic ML concepts and methods is the first in a series of articles illustrating ML algorithms applied to clinical datasets of older adults with cancer. The goal of this series is to encourage the discussion of these methods as possible adjuncts to traditional regression models, and in turn to increase understanding of the data most crucial for decision-making for older adults with cancer and their care teams.
Funding
Dr. Ramsdale received grants from the National Cancer Institute (K08CA248721) and the National Institutes on Aging (R03AG067977) to support this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
The authors report no conflicts of interest.
References
- 1.scikit-learn: Machine Learning in Python. (Accessed September 12, 2020, at https://scikit-learn.org/stable/.)
- 2.CRAN Task View: Machine Learning & Statistical Learning. (Accessed September 12, 2020, at https://cran.r-project.org/web/views/MachineLearning.html.)
- 3.SAS Visual Data Mining and Machine Learning Features. (Accessed September 12, 2020, at https://www.sas.com/en_us/software/visual-data-mining-machine-learning/features-list.html.)
- 4.User’s corner: Machine Learning. (Accessed September 12, 2020, at https://www.stata.com/stata-news/news33-4/users-corner/.)
- 5.Machine Learning: Latest Features. (Accessed September 12, 2020, at mathworks.com/solutions/machine-learning/features.html.)
- 6.Witt DR, Kellogg RA, Snyder MP, Dunn J. Windows into human health through wearables data analytics. Current Opinion in Biomedical Engineering 2019;9:28–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awoyemi JO, Adetunmbi AO, Oluwadare SA. Credit card fraud detection using machine learning techniques: A comparative analysis. 2017 International Conference on Computing Networking and Informatics (ICCNI); 2017 29–31 October. 2017. p. 1–9. [Google Scholar]
- 8.Kaneko Y, Yada K. A Deep Learning Approach for the Prediction of Retail Store Sales. 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW); 2016 12–15 December. 2016. p. 531–7. [Google Scholar]
- 9.Smith BD, Smith GL, Hurria A, Hortobagyi GN, Buchholz TA. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol 2009;27:2758–65. [DOI] [PubMed] [Google Scholar]
- 10.Singh H, Kanapuru B, Smith C, et al. FDA analysis of enrollment of older adults in clinical trials for cancer drug registration: A 10-year experience by the U.S. Food and Drug Administration. Journal of Clinical Oncology 2017;35:10009-. [Google Scholar]
- 11.Berry SD, Ngo L, Samelson EJ, Kiel DP. Competing Risk of Death: An Important Consideration in Studies of Older Adults. Journal of the American Geriatrics Society 2010;58:783–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scotté F, Bossi P, Carola E, et al. Addressing the quality of life needs of older patients with cancer: a SIOG consensus paper and practical guide. Annals of Oncology 2018;29:1718–26. [DOI] [PubMed] [Google Scholar]
- 13.Beaulieu-Jones BK, Orzechowski P, Moore JH. Mapping Patient Trajectories using Longitudinal Extraction and Deep Learning in the MIMIC-III Critical Care Database. Pac Symp Biocomput 2018;23:123–32. [PubMed] [Google Scholar]
- 14.Weisenthal SJ, Quill C, Farooq S, Kautz H, Zand MS. Predicting acute kidney injury at hospital re-entry using high-dimensional electronic health record data. PLoS One 2018;13:e0204920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg A, Fucile C, White RJ, et al. Visualizing nationwide variation in medicare Part D prescribing patterns. BMC Med Inform Decis Mak 2018;18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi LM, Myers TG, Fan Y, et al. Mining the National Cancer Institute Anticancer Drug Discovery Database: Cluster Analysis of Ellipticine Analogs with p53-Inverse and Central Nervous System-Selective Patterns of Activity. Molecular Pharmacology 1998;53:241. [DOI] [PubMed] [Google Scholar]
- 17.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akamine Y, Ueda Y, Ueno Y, et al. Application of hierarchical clustering to multi-parametric MR in prostate: Differentiation of tumor and normal tissue with high accuracy. Magn Reson Imaging 2020. [DOI] [PubMed] [Google Scholar]
- 19.Xu H Using machine learning to identify older adults at high risk for hospitalization and mortality via the Patient-Reported Outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE). In: Xu Huiwen SGMMAFLJPMMEERAPSJLJVGVKKBH, editor.2020; ASCO Quality Care Symposium: American Society of Clinical Oncology. [Google Scholar]
- 20.Soto Perez De Celis E, Li D, Sun C-L, et al. Patient-defined goals and preferences among older adults with cancer starting chemotherapy (CT). Journal of Clinical Oncology 2018;36:10009-. [Google Scholar]
- 21.van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Medical Research Methodology 2014;14:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohile SG, Dale W, Somerfield MR, Hurria A. Practical Assessment and Management of Vulnerabilities in Older Patients Receiving Chemotherapy: ASCO Guideline for Geriatric Oncology Summary. J Oncol Pract 2018;14:442–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wang D, Yao Z, et al. Machine Learning Models for Multiparametric Glioma Grading With Quantitative Result Interpretations. Frontiers in Neuroscience 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magesh P, Myloth R, Tom R. An Explainable Machine Learning Model for Early Detection of Parkinson’s Disease using LIME on DaTscan Imagery 2020. [DOI] [PubMed]
- 25.O’Neil C Weapons of Math Destruction: How Big Data Increases Inequality and Threatens Democracy: Crown Publishing Group; 2016.
- 26.Huesch MD, Mosher TJ. Using It or Losing It? The Case for Data Scientists Inside Health Care. NEJM Catalyst Innovations in Care Delivery 2017;Published online at: https://catalyst.nejm.org/doi/full/10.1056/CAT.17.0493. [Google Scholar]
- 27.Ming C, Viassolo V, Probst-Hensch N, Dinov ID, Chappuis PO, Katapodi MC. Machine learning-based lifetime breast cancer risk reclassification compared with the BOADICEA model: impact on screening recommendations. Br J Cancer 2020;123:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R. A Deep Learning Mammography-based Model for Improved Breast Cancer Risk Prediction. Radiology 2019;292:60–6. [DOI] [PubMed] [Google Scholar]
- 29.Nartowt BJ, Hart GR, Roffman DA, et al. Scoring colorectal cancer risk with an artificial neural network based on self-reportable personal health data. PLoS One 2019;14:e0221421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera M, Mirchandani R, Papa N, et al. PSA-based machine learning model improves prostate cancer risk stratification in a screening population. World J Urol 2020. [DOI] [PubMed] [Google Scholar]
- 31.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada M, Saito Y, Imaoka H, et al. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep 2019;9:14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschandl P, Codella N, Akay BN, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol 2019;20:938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama S, Hamada T, Higashi M, et al. Predicted Prognosis of Patients with Pancreatic Cancer by Machine Learning. Clin Cancer Res 2020;26:2411–21. [DOI] [PubMed] [Google Scholar]
- 35.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 2007;356:11–20. [DOI] [PubMed] [Google Scholar]
- 36.Wang HY, Sun BY, Zhu ZH, et al. Eight-signature classifier for prediction of nasopharyngeal [corrected] carcinoma survival. J Clin Oncol 2011;29:4516–25. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Xie J, Han Z, et al. Immunomarker Support Vector Machine Classifier for Prediction of Gastric Cancer Survival and Adjuvant Chemotherapeutic Benefit. Clin Cancer Res 2018;24:5574–84. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z, Zhan X, Xiang S, et al. SALMON: Survival Analysis Learning With Multi-Omics Neural Networks on Breast Cancer. Front Genet 2019;10:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton EA, Pujol TA, McDonald JF, Qiu P. Leveraging TCGA gene expression data to build predictive models for cancer drug response. BMC Bioinformatics 2020;21:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mucaki EJ, Zhao JZL, Lizotte DJ, Rogan PK. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduct Target Ther 2019;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat 2019;173:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C, Mezencev R, McDonald JF, Vannberg F. Open source machine-learning algorithms for the prediction of optimal cancer drug therapies. PLoS One 2017;12:e0186906. [DOI] [PMC free article] [PubMed] [Google Scholar]


