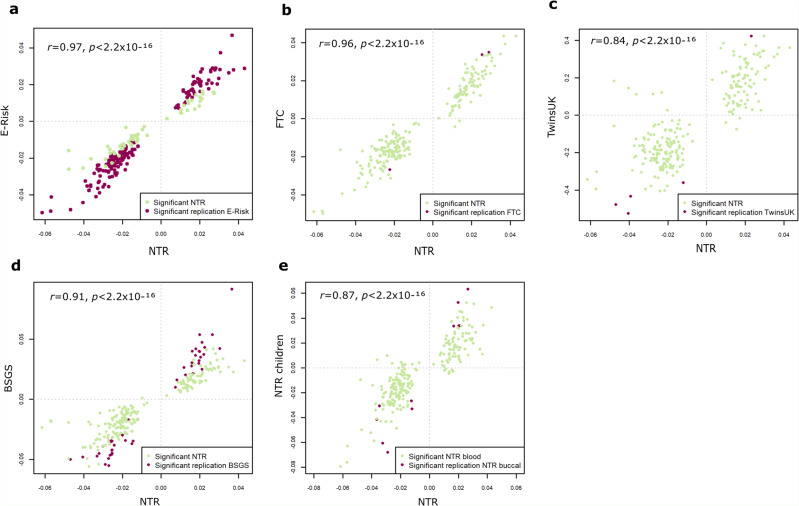
Fig. 1. Replication of MZ-DMPs identified in NTR in blood DNA methylation data from four independent twin cohorts and buccal DNA methylation data from one independent twin cohort.
Scatterplots showing the estimates (methylation beta-value difference between MZ twins and controls; a positive difference corresponds to a higher methylation level in MZ twins) in the discovery and replication cohorts for MZ-DMPs identified in NTR. The x axis shows the estimates in the discovery EWAS in the Netherlands Twin Register (NTR, N = 1957, whole blood). The y-axis shows the estimates in a The Environmental Risk Longitudinal Twin Study (E-Risk, N = 1164, whole blood). b The Finnish Twin Cohort (FTC, N = 1708, whole blood). c The UK Adult Twin Registry (TwinsUK, N = 492, whole blood). In TwinsUK, residuals obtained after correcting for covariates were analyzed instead of methylation beta values. d The Brisbane Systems Genetic Study (BSGS, N = 356, whole blood). e An independent child data set from the NTR (N = 765, buccal). Each dot is one methylation site. Methylation sites that replicate following stringent Bonferroni correction for 243 tests and after correction for inflation of genome-wide test statistics, where applicable, are displayed in dark purple, all other sites are shown in green. r = correlation.