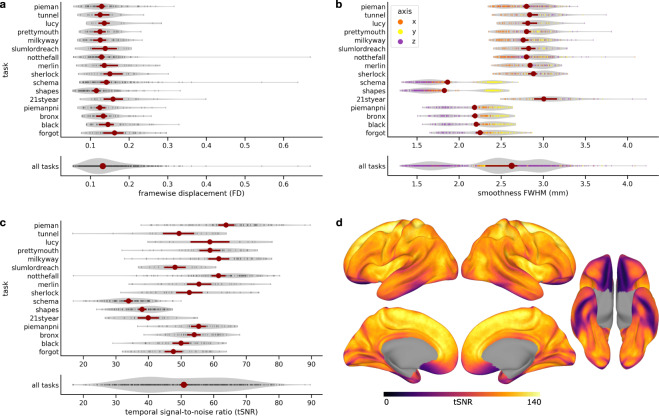
Fig. 2.
Data quality summarized in terms of head motion, spatial smoothness, and temporal signal-to-noise ratio (tSNR). (a) Median framewise displacement (FD) summarizes head motion. Each black tick mark represents the median FD (x-axis) for a given scanning run and subject for each task (i.e. story; y-axis). Violin plots illustrate the distribution of median FDs across all runs and subjects for a given task. The red markers indicate the median FD across all runs for a given task, and the red error bars indicate the 95% bootstrap confidence interval for the median based on randomly sampling runs with replacement. At bottom, FD is summarized across all tasks. (b) Spatial smoothness is summarized using AFNI’s FWHM smoothness for each scanning run across each task. Spatial smoothness was computed in volumetric subject-specific functional (EPI) space using subject-specific whole-brain masks with minimal preprocessing (realignment and susceptibility distortion in fMRIPrep, detrending in AFNI). Each tick mark represents the spatial smoothness for a given run in a given acquisition axis (orange: x-axis, i.e. left–right; yellow: y-axis, i.e. anterior–posterior; red: z-axis, i.e. inferior–superior). Violin plots capture the distribution of smoothness estimates for x-, y-, and z-axes across all runs and subjects in a given task. The red markers indicate the median combined (i.e. geometric mean across x-, y-, and z-axes) smoothness across all runs and subjects for a given task (red error bars indicate 95% bootstrap confidence interval of median). At bottom, smoothness is summarized across all tasks. The multiple lobes of the distribution reflect differing acquisition parameters used with the Skyra and Prisma scanner models. (c) tSNR maps were constructed by computing the voxelwise mean signal over time divided by the standard deviation over time. Each black tick mark represents the median of the tSNR map (x-axis) for a given scanner run and subject for each task (y-axis). Violin plots reflect the distribution of tSNR values across all runs and subjects for a given task. The red markers indicate the mean tSNR across all runs for a given task (red error bars indicate 95% bootstrap confidence interval of mean). At bottom, the tSNR is summarized across all tasks. The two lobes of the distribution reflect older pulse sequences with larger voxels (and higher tSNR) and newer multiband pulse sequences with smaller voxels (and lower tSNR). See the plot_qc.py script in the code/ directory for details. (d) Distribution of tSNR across cortex. The color bar reflects the median vertex-wise tSNR. Note that unlike panel c, here tSNR was computed in fsaverage6 space for vertex-wise summarization and visualization, which may inflate the tSNR values.