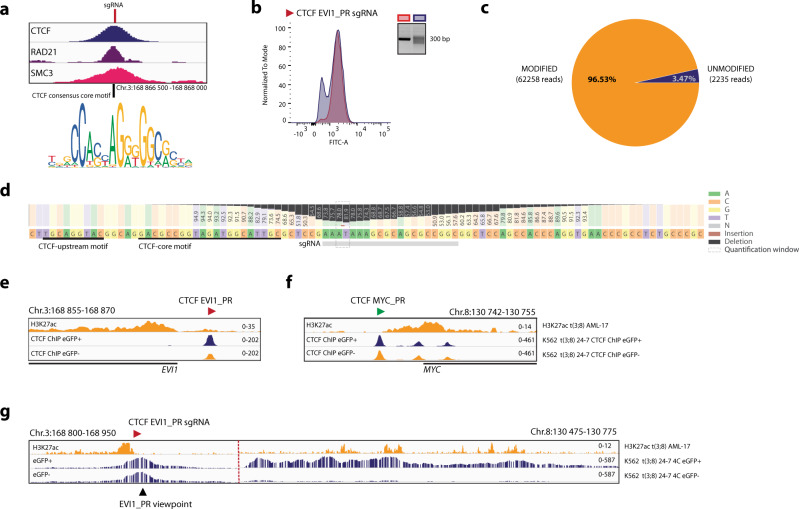
Fig. 6. CTCF binding site upstream of the EVI1 promoter hijacks a MYC SE in t(3;8) AML.
a ChIP-seq data of CTCF (blue) and the cohesin subunits RAD21 (purple) and SMC3 (pink) in K562, with a zoom-in on the EVI1 promoter binding site. The vertical line indicates the exact cleavage site of the sgRNA and the CTCF motif as described by JASPAR63 below. b Flow cytometry overlay plot after targeting the CTCF EVI1_PR binding site by sgRNA (clone 8-4, blue graph) and in red the control cells (clone 8-4, no Cas9). On the right of the graph, the mutations introduced by the single sgRNA in the amplicon over the cutting site are shown by PCR. Source data are provided as a Source Data file. c Amplicon-seq data showing the percentage of modified (orange) and unmodified (blue) reads in the eGFP− sorted cell fraction after targeting CTCF EVI1_PR. d Amplicon-seq data showing the mutations in the nucleotides around the Cas9 cleavage site, in the eGFP− sorted cell fraction after targeting CTCF EVI1_PR with sgRNA. The bars and numbers indicate the percentage of reads found with the particular mutation, below the locations of the sgRNA (gray bar) and the CTCF motifs (black lines). e CTCF ChIP-seq presenting CTCF occupancy in the eGFP+ (clone 24-7, blue), and eGFP− (clone 24-7, orange) fractions after targeting CTCF EVI1_PR with the sgRNA. The top H3K27ac track (orange) indicates the presence of an active promoter. f The same CTCF ChIP-seq tracks as in e are shown, but here presenting unchanged CTCF occupancy at the CTCF binding site upstream of the MYC promoter. g Chromatin interaction at the MYC SE for eGFP+ and eGFP− cells (clone 24-7), shown by 4C-seq with the EVI1 promoter as viewpoint after targeting the CTCF motif with the sgRNA.