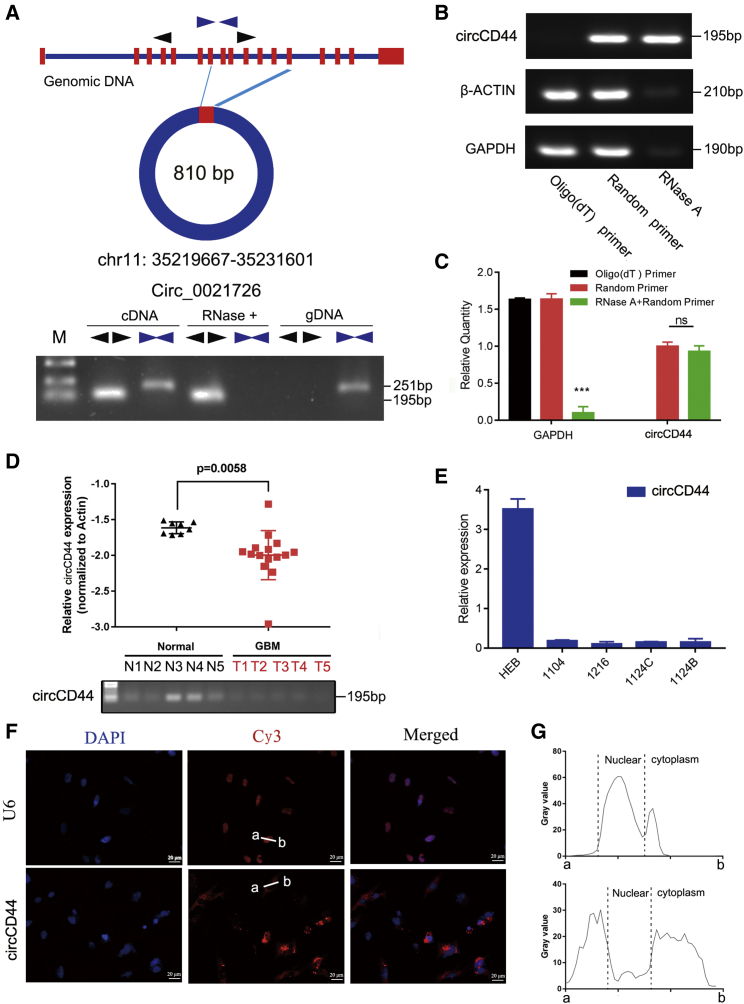
Figure 3.
circCD44 is a circular RNA and is downregulated in GBM
(A) Schematic representation of circCD44 formation (top). The RNAs were detected by PCR. Divergent primers could produce circRNAs in cDNA but not in genomic DNA (gDNA); convergent primers could produce cDNA and gDNA (bottom). (B) PCR and RT-qPCR showed that circCD44 cannot be reverse-transcribed using Olig(dt) primer, while it can be reverse-transcribed using random primers. (C) RT-qPCR showed that circCD44 resists degradation of RNase R, while gapdh mRNA can be degraded by RNase R. (D) The expression level of circCD44 was detected by RT-qPCR in GBM tissues (n = 14) and normal brain tissues (n = 8). (E) The expression level of circCD44 was detected by RT-qPCR in a human normal glial cell line (HEB) and primary glioma cells (1104, 1216, 1124C, 1124B). (F) Confocal FISH was performed to determine the location of circCD44 in 1124C cells. Scale bars, 20 μm. (G) Fluorescence signals were measured with ImageJ software. ∗p<0.05; ∗∗p<0.01:∗∗∗p<0.001.