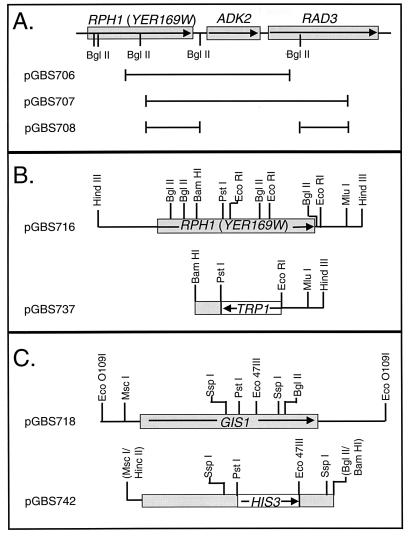
FIG. 1.
Restriction maps of yeast chromosomal inserts in selected plasmids used in cloning and disruption of YER169w (RPH1) and YDR096w (GIS1). Arrows indicate the direction of transcription of genes indicated by boxes. (A) Sketch of the region of chromosome V carrying YER169w and adjacent genes which were included in the GAL4AD-YER169w fusions that activated the URSPHR1 reporter constructs. The chromosomal DNA carried by plasmids pGBS706, pGBS707, and pGBS708 are indicated by the black lines beneath the map. (B) Restriction map of the yeast chromosomal DNA fragments carried by pGBS716 and by the derivative plasmid pGBS737 which was used to disrupt YER169w. (C) Restriction map of the 4.6-kbp chromosomal DNA fragment carried by pGBS718 and of the gene disruption in plasmid pGBS742. Among the SspI and Eco47III sites in the fragments, only those sites used in subcloning and directed homologous recombination are shown. Restriction sites in parentheses were lost during subcloning.