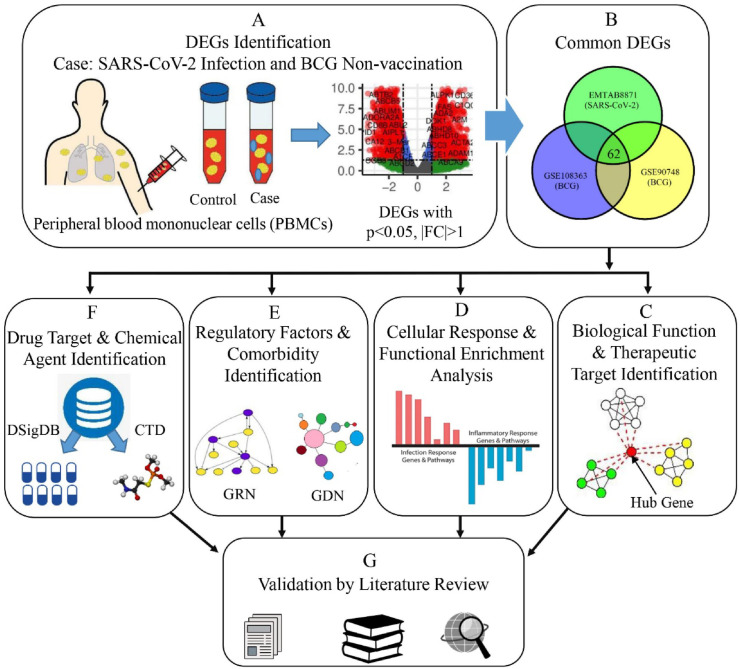
Fig. 1.
Schematic diagram outlining the workflow of our proposed approach. (A) To conduct differential expression analysis, we have designed three individual experiments for each of the datasets. In those experiments, the case conditions were SARS-CoV-2 infection and BCG non-vaccination (2 datasets), and the control conditions were healthy status and BCG vaccination (2 datasets) respectively. (B) Common DEGs were then identified for both health conditions. (C) Biological functions of these DEGs were assessed and therapeutic targets were found by PPI analysis. (D) Functional enrichment analysis was performed with GO and cell signalling pathway databases. (E) Regulatory elements and possible comorbidities were determined. (F) Putative drug candidates and chemical agents were identified using curated databases. (G) All the gained results were validated through an extensive literature review.