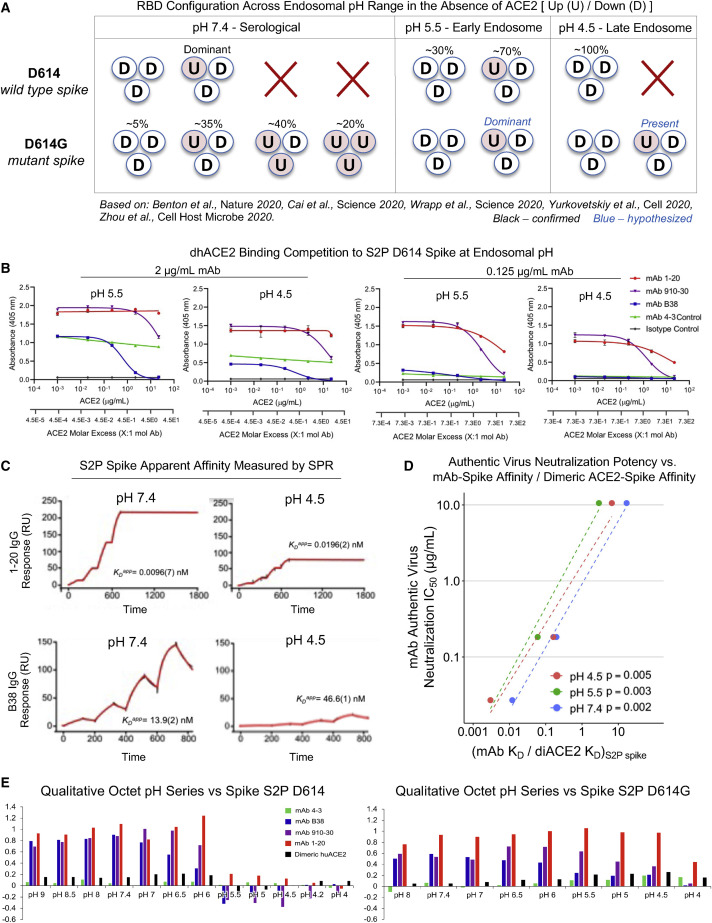
Figure 4.
Up/down conformational changes of RBD influence IGHV3-53/3-56 antibody class recognition of spike protein across the serological to endosomal pH range
(A) Schematic of RBD conformational states inferred by cryo-EM and experimental analysis for un-ligated D614 and D614G spike. U and D denote up and down RBD configurations, respectively. Percentages denote observed particle populations.
(B) dhACE2 competition ELISA at endosomal pH against SARS-CoV-2 S2P D614 protein. dhACE2 concentration is provided as both μg/mL and ACE2 molar excess units. Data are represented as means ± SDs.
(C) Single-cycle SPR kinetic assays for 1-20 and B38 IgG at serological and endosomal pH against biotinylated spike. Black traces represent experimental data and red traces represent the fit to a 1:1 interaction model. The number in parentheses represents the error of the fit in the last digit.
(D) Correlations between authentic virus neutralization potency (Figure 2D) versus the ratio of antibody IgG affinity to spike S2P (Figure S4A) divided by dhACE2 affinity to spike S2P (reported from Zhou et al., 2020b).
(E) Qualitative octet pH series for wild-type S2P spike and escape mutant D614G S2P spike across a range of pH values.
See also Figures S3 and S4.