Abstract
Single neuron-specific drivers are important tools for visualizing neuron anatomy, manipulating neuron activity and gene rescue experiments. We report here that genomic regions upstream of the C. elegans bHLH-PAS gene hlh-34 can be used to drive gene expression exclusively in the AVH interneuron pair and not, as previously reported, the AVJ interneuron pair.
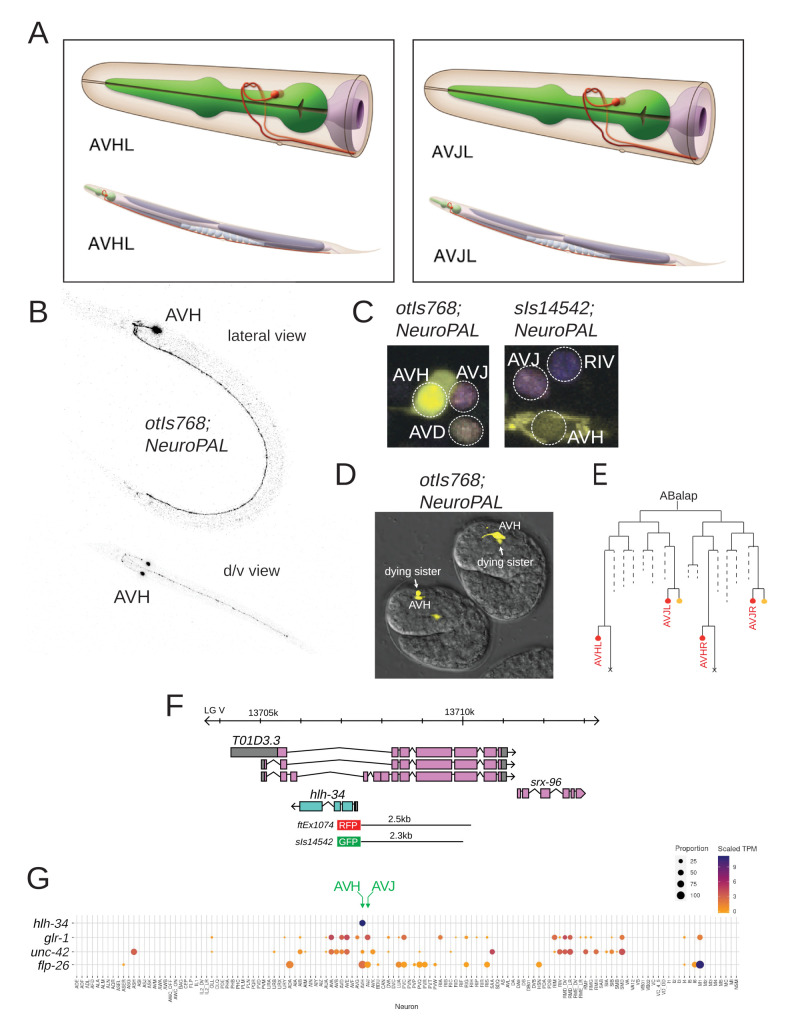
Figure 1. AVH and AVJ neurons and hlh-34 expression.
A: AVH and AVJ display a nearly identical position and axon trajectory in the lateral ganglion of C. elegans. Images used with permission from Wormatlas.
B: Whole-animal lateral and dorsoventral overviews of hlh-34prom::gfp (otIs768 transgene) expressing young larvae. Animals imaged in a NeuroPAL (otIs669) background with only GFP emission shown for simplicity.
C: Expression pattern of otIs768 and sIs14542 transgenic reporters in a NeuroPAL (otIs669) background. GFP signal is only observed in the AVH neuron.
D: Embryonic expression pattern of otIs768. At ~360 minutes the GFP signal is observed in the AVH neuron and its dying sibling cell. All animals were imaged in aNeuroPAL (otIs669) background with only GFP emission shown for simplicity.
E: Lineages of AVH/J in red. CEPsoDL/R shown in orange. Dying siblings of AVH shown as ‘x’.
F: hlh-34 locus and promoter fragment used in the sIs14542 transgene. Coordinates are V:13709947..13712813 in WS140 and V:13707915..13707382 in WS280. The size of the promoter used in the leEx1692 strain, kindly provided by Ian Hope, could not be unambiguously determined. The strain used in the first publication on hlh-34 (Cunningham et al. 2012) used 2.5kb upstream of hlh-34.
G: Heatmap of scRNA expression (Taylor et al. 2021), showing hlh-34 expression and other markers of AVH and AVJ identity.
Description
bHLH-PAS transcription factors are a metazoan-specific family of transcription factors with diverse functions within and outside the nervous system (Kewley et al. 2004). Caenorhabditis elegans contains five bHLH-PAS proteins: (1) ahr-1, an ortholog of the aryl hydrocarbon receptor(Powell-Coffman et al. 1998); (2) aha-1, an ortholog ofthe Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT), which is a common dimerization partner for many, but not all bHLH-PAS genes (Powell-Coffman et al. 1998);(3) hif-1, an ortholog of hypoxia-inducible factor HIF1alpha (Jiang et al. 2001);(4) CKY-1, which orthology prediction tools consider to be an ortholog of NPAS4, even though its bHLH domain is very degenerate (nevertheless, CKY-1 heterodimerizes, like other C. elegans bHLH-PAS proteins, with the common AHA-1 partner protein in yeast 2 hybrid assays (Grove et al. 2009)); (5) hlh-34, which Marvvel (Wang et al. 2019) and Ortholist (Shaye and Greenwald 2011) predict to be an ortholog of both the NPAS1/3 and the SIM1/2 subgroups of bHLH-PAS proteins (Yan et al. 2014).However,HLH-34 contains neither of the domains that are found, in addition to the canonical bHLH and PAS domains, in either vertebrate NPAS (PAS_11 domain) or SIM (SIM_C) proteins. HLH-34 may therefore reflect an ancestral version of both subgroups.
In this paper, we re-assess the previously reported expression pattern of the hlh-34 gene. An intriguing previous study that analyzed feeding control in C. elegans used 2.5kb of the hlh-34 promoter in a reporter gene construct to assess expression of the gene(Cunningham et al. 2012). This reporter was described to be expressed exclusively in a single neuron pair that the authors tentatively identified as the AVJ neurons (Cunningham et al. 2012). Based on rescue experiments that the authors performed with the hlh-34 promoter, feeding behavior functions were ascribed to AVJ (Cunningham et al. 2012). Moreover, again based on the expression assignment of the hlh-34 promoter, an interesting recent study implicated the AVJ neurons in cold tolerance behavior (Takagaki et al. 2020). Lastly, the neurogenin homolog ngn-1 was recently reported to function in the AVJ neurons, based on its regulation of hlh-34 reporter expression (Christensen et al. 2020).
However, AVJ and its neighboring AVH neuron pair are two bilaterally symmetric neuron pairs that are notoriously difficult to distinguish based on their almost identical cell body position and neurite trajectory (White et al. 1986)(Fig.1A).
In the context of studying fate specification of the AVJ and AVH neurons (Berghoff et al. 2021), we obtained an hlh-34 promoter-based reporter line from Ian Hope, leEx1692, established in initial attempts to broadly studying gene expression (Hope et al. 1996). We found that this transgene is expressed in a single head neuron pair, extending its neurite along the ventral nerve cord (Fig.1B), consistent with this neuron being either the AVJ or AVH neuron pair. After chromosomal integration, we crossed this transgene (otIs768) with NeuroPAL, a transgene that provides a multicolor bar code for all neurons in the C. elegans nervous system, with distinct color codes for AVH and AVJ (Yemini et al. 2021). We find that the hlh-34prom::gfp signal from the otIs768 transgene overlaps with the AVH and not the AVJ signal(Fig.1C). We validated this assessment by crossing otIs768 with an AVH-expressed marker that is not in AVJ (unc-42::rfp) and an AVJ expressed marker, that is not in AVH (glr-1prom::rfp). We observed an overlap with unc-42:rfp but not glr-1prom::rfp. We also obtained another hlh-34 promoter gfp reporter from the Moerman & Baillie groups, sIs14542 (Hunt-Newbury et al. 2007) (Fig.1F).Using NeuroPAL, we confirmed sIs14542 expression in AVH, but not AVJ (Fig.1C).
We further corroborated hlh-34prom expression in AVH by observing that in the embryo, expression of otIs768 is first transiently observed in 4 cells, two of which show signs of cell death (Fig.1D); later in embryogenesis, and then during all stages of postembryonic and adult development, expression becomes restricted to 2 cells. This is consistent with expression in the bilateral AVH neuron pair, since their two sisters cells are, unlike the sisters of the AVJ neuron pair, destined to die by apotosis (Sulston et al. 1983) (Fig.1E).
Lastly, single cell RNA profiling of the entire C. elegans nervous system reveals strong and selective expression of hlh-34 transcripts exclusively in the AVH neuron (Taylor et al. 2021)(Fig.1G). The scRNA data not only confirm hlh-34 expression in AVH but also corroborates the expression of the markers that we used to distinguish AVH from AVJ (Fig.1G). Since all previously described hlh-34 reporter constructs only encapsulate parts of the 5’ region of the gene and because the gene has an unusual location in the intron of another gene (Fig.1F), the scRNA data represent an important, independent validation of the assignment of the existing reporter gene patterns to AVH and not AVJ.
The expression of the hlh-34 promoter in AVH rather than AVJ indicates that functions previously ascribed to the AVJ neuron need to be re-ascribed to the AVH neuron. This applies to the above-mentioned feeding behavior function initially ascribed to AVJ (Cunningham et al. 2012) which now has to be assigned to AVH. Similarly, the implication of AVJ in cold tolerance behavior, based on the expression overlap of a gene involved in cold tolerance, xdh-1, and hlh-34prom::rfp, and based on the rescue of the xdh-1 mutant phenotype with an hlh-34prom driver (Takagaki et al. 2020), needs to be re-assigned to the AVH neuron. Lastly, the function of the ngn-1 bHLH gene of regulating hlh-34 (Christensen et al. 2020) needs to be re-assigned to AVH.
In conclusion, the 5’ region of the hlh-34 gene is a useful tool to gain genetic access to the AVH neuron.
Methods
Strains were examined with a laser scanning LSM 880 microscope.
Reagents
The following transgenes were used:
OH16483: unc-119(ed3); otIs768[hlh-34prom::gfp, unc-119(+)]
BC15839: dpy-5(e907); sIs14542[hlh-34prom::gfp, dpy-5(+)]
OH15363: otIs669[NeuroPAL]; him-5(e1490)
Acknowledgments
Acknowledgments
We thank Kaveh Ashrafi and Ian Hope for sending us the hlh-34 reporter strains and to Wormbase for help with identifying promoter coordinates.
Funding
HHMI
References
- Berghoff EG, Glenwinkel L, Bhattacharya A, Sun H, Varol E, Mohammadi N, Antone A, Feng Y, Nguyen K, Cook SJ, Wood JF, Masoudi N, Cros CC, Ramadan YH, Ferkey DM, Hall DH, Hobert O. The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. Elife. 2021 Jun 24;10 doi: 10.7554/eLife.64903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen EL, Beasley A, Radchuk J, Mielko ZE, Preston E, Stuckett S, Murray JI, Hudson ML. ngn-1/neurogenin Activates Transcription of Multiple Terminal Selector Transcription Factors in the Caenorhabditis elegans Nervous System. G3 (Bethesda) 2020 Jun 01;10(6):1949–1962. doi: 10.1534/g3.120.401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KA, Hua Z, Srinivasan S, Liu J, Lee BH, Edwards RH, Ashrafi K. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 2012 Jul 01;16(1):113–121. doi: 10.1016/j.cmet.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009 Jul 23;138(2):314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Albertson DG, Martinelli SD, Lynch AS, Sonnhammer E, Durbin R. The C. elegans expression pattern database: a beginning. Trends Genet. 1996 Sep 01;12(9):370–371. [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007 Sep 01;5(9):e237–e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A. 2001 Jun 26;98(14):7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004 Feb 01;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci U S A. 1998 Mar 17;95(6):2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011 May 25;6(5):e20085–e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983 Nov 01;100(1):64–6119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Takagaki N, Ohta A, Ohnishi K, Kawanabe A, Minakuchi Y, Toyoda A, Fujiwara Y, Kuhara A. The mechanoreceptor DEG-1 regulates cold tolerance in Caenorhabditis elegans. EMBO Rep. 2020 Feb 01;21(3):e48671–e48671. doi: 10.15252/embr.201948671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, Poff A, Basavaraju M, Rafi I, Yemini E, Cook SJ, Abrams A, Vidal B, Cros C, Tavazoie S, Sestan N, Hammarlund M, Hobert O, Miller DM 3rd. Molecular topography of an entire nervous system. Cell. 2021 Jul 01;184(16):4329–4347.e23. doi: 10.1016/j.cell.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Al-Ouran R, Hu Y, Kim SY, Wan YW, Wangler MF, Yamamoto S, Chao HT, Comjean A, Mohr SE, UDN. Perrimon N, Liu Z, Bellen HJ. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am J Hum Genet. 2017 May 11;100(6):843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976 Aug 10;275(938):327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- Yan J, Ma Z, Xu X, Guo AY. Evolution, functional divergence and conserved exon-intron structure of bHLH/PAS gene family. Mol Genet Genomics. 2013 Nov 01;289(1):25–36. doi: 10.1007/s00438-013-0786-0. [DOI] [PubMed] [Google Scholar]
- Yemini E, Lin A, Nejatbakhsh A, Varol E, Sun R, Mena GE, Samuel ADT, Paninski L, Venkatachalam V, Hobert O. NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans. Cell. 2020 Dec 29;184(1):272–288.e11. doi: 10.1016/j.cell.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]