Fig. 1.
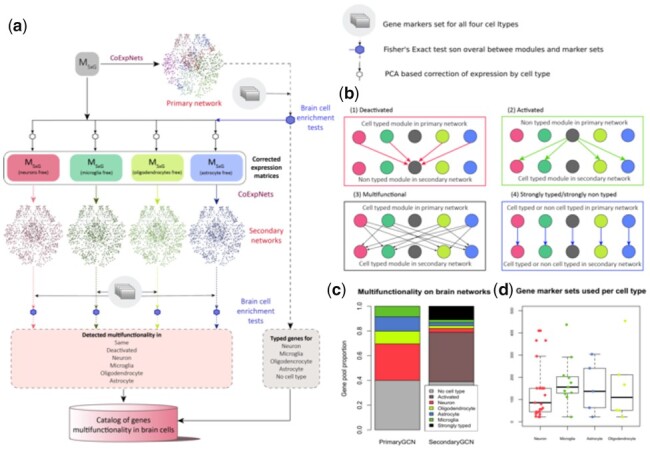
(a) GMSCA generates a list of triplets <gene, cell type, function> for the genes included in the initial gene expression profiling matrix, MSxG. First, a WGCNA+k-means co-expression network called the primary network is created. Its modules are then tested (Fisher’s Exact Test, FET) for enrichment of cell type markers, in this case with brain cell type marker sets for neurons (in red), microglia (in green), oligodendrocytes (in light green) and astrocytes (in blue). Those modules with a clear signal (FET P < 0.05 on just a single cell type) are selected and their corresponding cell signal removed (see Section 2) from expression to generate a new M’SxG. GMSCA creates a new co-expression network (these are called secondary) for each expression matrix and annotates their modules in the same way. Cell-type enriched modules in both primary and secondary networks generate as many triplets (gene, cell type, function) as genes in the module. (b) Any gene found in a cell type enriched module is tagged by GMSCA as ‘typed’. When a gene in a primary co-expression network shifts from a cell-type enriched module to a non-cell-type enriched module in the secondary co-expression network, we say the gene is deactivated (red arrows). When it goes from a non-cell-type enriched module to a cell-type enriched module, we say the gene is activated (green arrows). A gene is multifunctional when it goes from a cell-type enriched module to a module with a different cell type enrichment (black arrows). A gene is strongly typed when it goes from a cell-type enriched module to another module with the same cell-type enrichment. It is strongly non-typed when the primary module and secondary module are both non-cell-type enriched. (c) An average primary co-expression network tags 37% of genes as non-typed, another 30% as neuronal, 10% as oligodendrocytic, 13% as astrocytic and 8% as microglial. In a secondary network, 37% of non-typed genes become typed and 42.5% of typed genes become deactivated. Also, as 9.1% of the genes are tagged as pleiotropic in a single network, we can gain up to 36.4% annotations of pleiotropy with all four secondary networks GMSCA creates in this case. (d) Number and size of gene markers set used by GMSCA for each cell type, in this article
