Figure 1.
Germ-free mice with increased functional connectivity at the whole-brain level
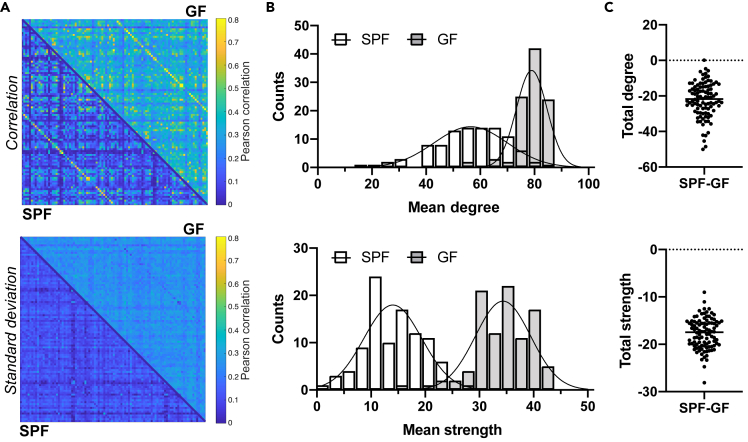
(A) Top: The Pearson correlation values expressing functional connectivity strength between pairs of anatomically defined nodes were expressed in an adjacency matrix for all 98 × 98 connections, with the SPF group in the lower triangle of the matrix and in the GF group in upper triangle of the matrix. Bottom: the corresponding standard deviation values of the group-wise Pearson correlation values are given. Color bar represents the correlation range from 0 (dark blue) to 0.8 (yellow).
(B) Degree and node strength distribution for SPF and GF group (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.001).
(C) Difference in total degree and node strength between SPF and GF mice (Student's t test, p < 0.001).
Data are represented as a split adjacency matrix (average Pearson correlation coefficient across animals per connection and related SD), histogram (counts of mean degree or mean node strength values), and individual data points. SPF n = 8, GF n = 15.