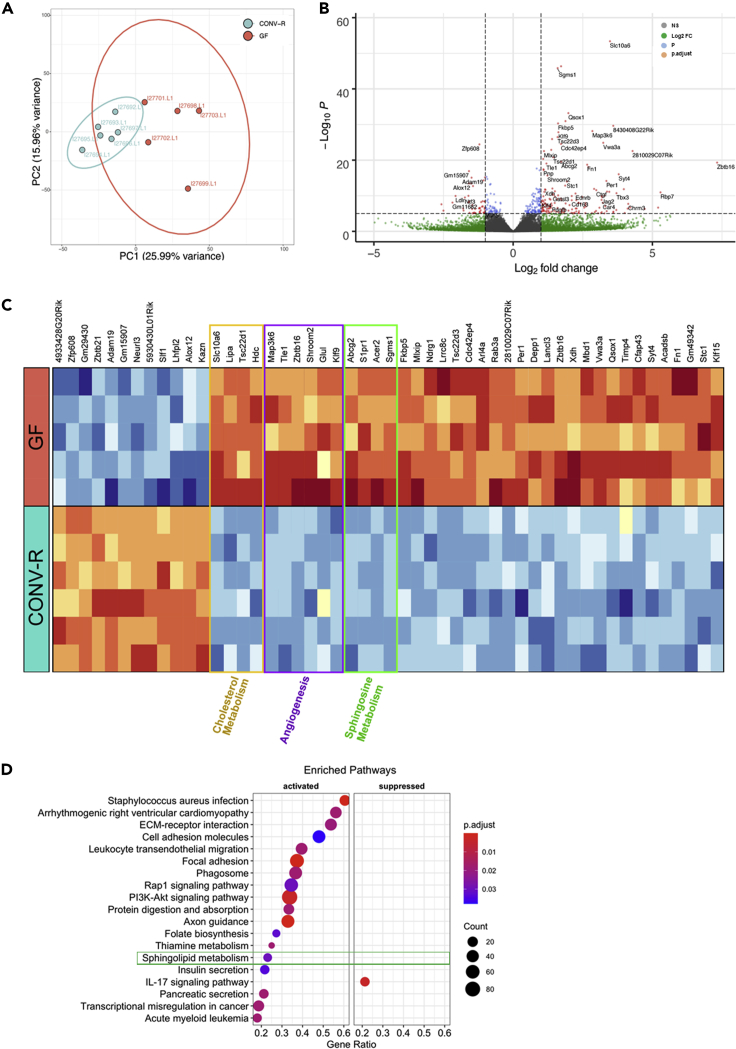
Figure 2.
Comparative analysis of RNA-sequencing data of isolated liver sinusoidal endothelial cells of germ-free (GF) vs. conventionally raised (CONV-R) C57BL/6J mice
(A) Principle component analysis (PCA) of the sequenced GF (red, n = 5) vs CONV-R (turquoise, n = 6) mice. PC1 shows a variance of 25.99% and PC2 of 15.96%.
(B) Volcano Plot of differentially expressed genes (DEGs). The genes are color coded according to difference in expression (Log2 fold change, Log2FC) and significance (adjusted p value, p.adjust). Gray: no difference in expression; not significant. Green: difference in expression, but not significant. Blue: significant difference, but the expression change is within [−1,1] Red: significant difference and high range expression change.
(C) Heatmap of the top 50 differentially expressed genes in hepatic endothelium comparing GF with CONV-R mice. The gold, violet and green frames highlight DEGs involved in cholesterol metabolism, angiogenesis and sphingosine metabolism, respectively.
(D) Gene set enrichment analysis of KEGG pathways. 19 KEGG pathways were significantly enriched at GF housing conditions. Pathways with a positive enrichment score were considered to be activated. The size of the dots corresponds to the number of genes in the reference gene set. The gene ratio is equal to the number of genes in the leading-edge subset divided by the total size of the gene set. The color of the dots corresponds to the adjusted p value (p.adjust; Benjamini-Hochberg method). See also Figures S1, S2, S4, and Table S1.