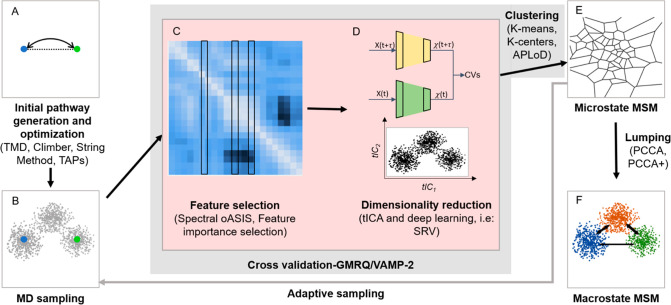
Figure 1.
Key steps in MSM construction for studying functional conformational changes in proteins. (A) Pathways between two or more end points of the functional conformational changes are generated and optimized to obtain minimum free energy pathways. (B) Extensive MD simulations are performed starting from these pathways. (C) Several relevant features or physical coordinates are selected. (D) Dimensionality reduction is performed using the selected features as input. (E) Reduced dimension data is discretized to obtain microstates and the MSM is estimated. (F) Kinetic lumping is performed to group microstates to macrostates.