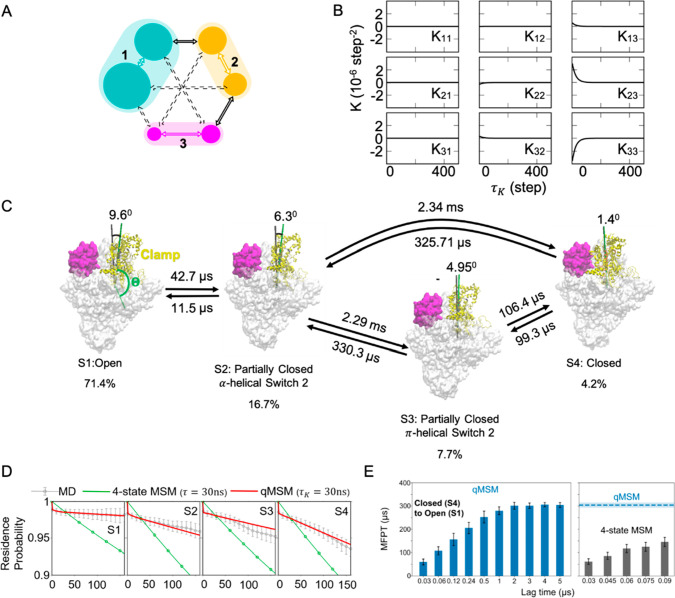
Figure 5.
qMSMs afford precise models with a handful of states. (A) Schematic of a simple three-state model. (B) Memory kernel tensor (K) of the three-state model. (C) The mechanism of the bacterial RNA polymerase clamp domain opening is shown, where four macrostates and the MFPTs between them are identified and estimated by the qMSM. (D) Chapman–Kolmogorov tests of the qMSM and four-state MSM are compared to MD simulations. (E) MFPTs from S4 to S1 estimated using the qMSM (left) and four-state MSM (right) are shown as a function of lag time. Panels (A) and (B) are reproduced with permission from ref (43). AIP Publishing, 2020. Panels (C)–(E) are adapted with permission from ref (90). National Academy of Sciences, 2021.