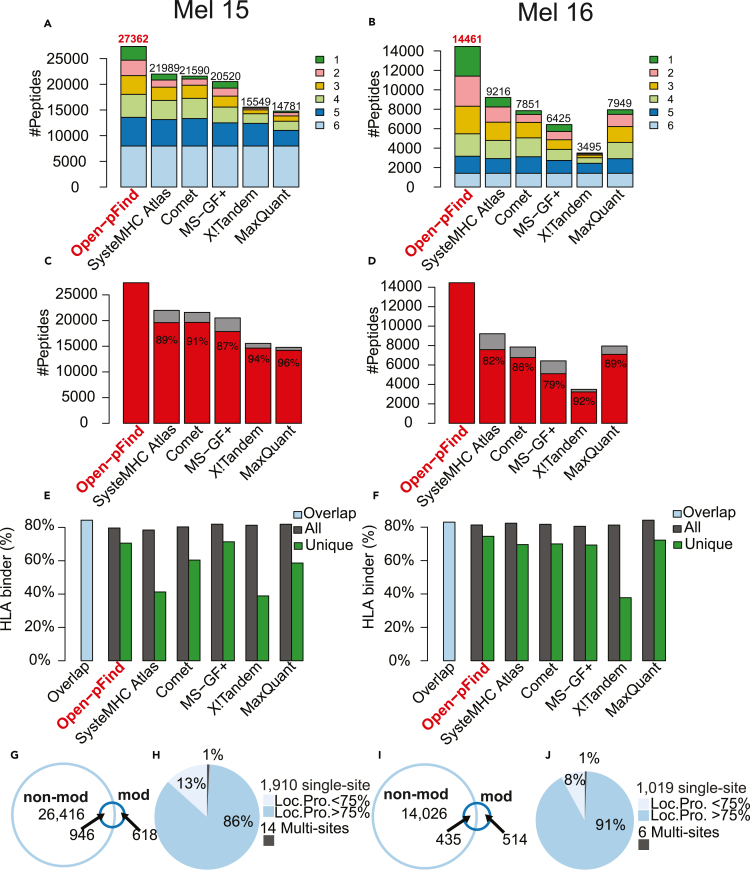
Figure 1.
Search engine comparison using HLA class I data from two melanoma cell lines (Left column: Mel15 and Right column: Mel16), see also Table S1
(A and B) The total number of non-modified peptides (length between 8 and 12) identified by each method, with different colors indicating the number of methods supporting a particular peptide identification.
(C and D) The percentage of non-modified peptides covered by open-pFind identifications for each method.
(E and F) HLA-binder percentages for the overlapping identifications of the six methods (blue bar) and for the total (black bar) and unique (green bar) identifications of each method, respectively.
(G and I) Venn diagrams comparing peptide sequences from modified (mod) and non-modified (non-mod) peptides.
(H and J) The percentages of peptides with single and multiple modification sites, with the singly modified sites further divided by localization probability (Loc. Pro.) computed by PTMiner.