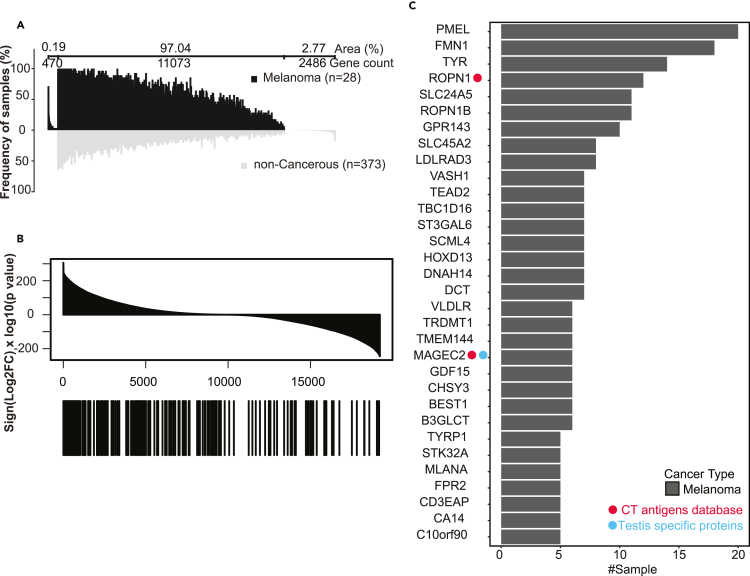
Figure 7.
Tumor associated antigen analysis for melanoma, see also Figures S6, S7, and Table S5
(A) Differential immunopeptidomic analysis of antigen source genes from melanoma HLA class I samples (n = 28) and comparative profiling to non-cancerous HLA class I samples (n = 373). Profiling is based on the detection frequency for individual genes. The numbers of genes that are unique to melanoma samples, unique to non-cancerous samples, and shared between melanoma and non-cancerous samples are indicated above the graph, together with the respective area under the curve in percentage of total area (i.e., the sum of sample frequency for each gene divided by that for all genes). Genes are sorted according to the sample numbers within each of the three groups.
(B) All genes quantified in both TCGA and GTEx studies were sorted by signed −log10(pValue) in the top panel. The locations of the 201 recurrently identified, melanoma-associated antigen source genes in the sorted list are annotated by the black bars in the bottom panel. p values were derived from Student’s t-test.
(C) Bar plot showing the number of tumor samples for each of the 32 melanoma-associated antigen source genes identified in more than five melanoma samples and with significantly higher expression in the TCGA melanoma samples compared to GTEx normal skin tissues. The red and blue dots on the left indicate genes from the CT antigens database and the GTEx-supported testis-specific proteins, respectively.