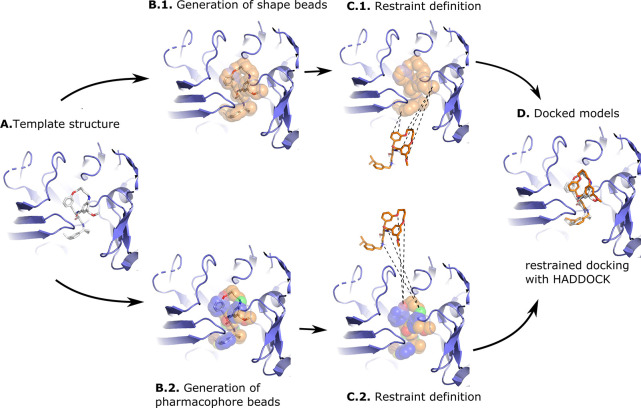
Figure 1.
Illustration of the shape-based and the pharmacophore-based docking protocol. Panel (A) shows a suitable receptor template identified based on the similarity of its bound ligand and the ligand to be docked (see Materials and Methods). Panel (B) shows the heavy atoms of the crystallographic compound transformed into shape beads (B.1) or pharmacophore beads with the colored beads representing different pharmacophore properties (B.2). The crystallographic compound is then removed from the pocket, and restraints are defined between the beads and the conformers: In the shape-based protocol (C.1) restraints are defined between all atoms of the compound and all beads of the shape, while in the pharmacophore-based protocol (C.2), restraints are defined between atoms of the compound and beads that share identical pharmacophore features. Panel (D) shows a docked model superimposed onto the template structure. The protein receptor is shown as slate cartoon, the crystallographic compound as white sticks, the generated and docked compounds as orange sticks, and the shape beads as transparent orange spheres. All molecular graphics were generated with PyMOL.26