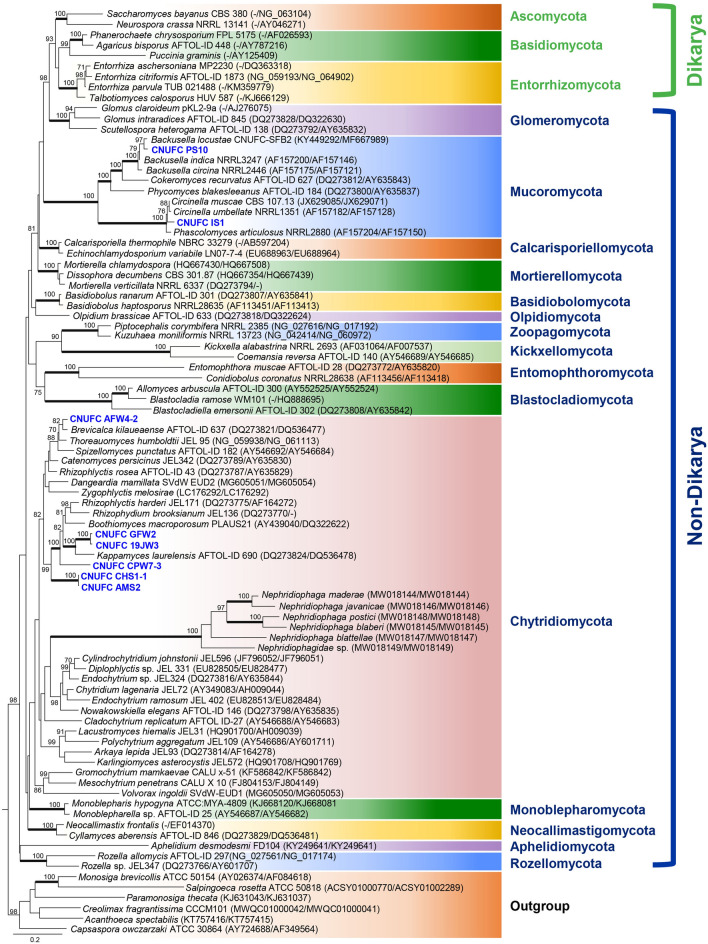
Fig. 2.
Phylogram generated from Maximum Likelihood analysis based on LSU and SSU combined sequence data showing evolutionary relationships of basal fungal phyla and relatives. The tree was inferred using 89 taxa and 3,790 aligned nucleotide sites. Support in nodes is indicated above or below branches and is represented by bootstrap values (ML analysis) of 70% and higher. Full-supported branches (100% BS) are highlighted by thickened branches. Analysis performed using RAxML-HPC2 on the CIPRES Science Gateway server with 1,000 bootstrap replicates and the GTRGAMMA model of nucleotide substitution. The tree includes six outgroup taxa including Acanthoeca spectabilis, Capsaspora owczarzaki, Creolimax fragrantissima, Monosiga brevicollis, Paramonosiga thecata and Salpingoeca rosetta. Strains in bold blue refer to taxa not designated yet to a specific group. The corresponding sequences were generated during this study