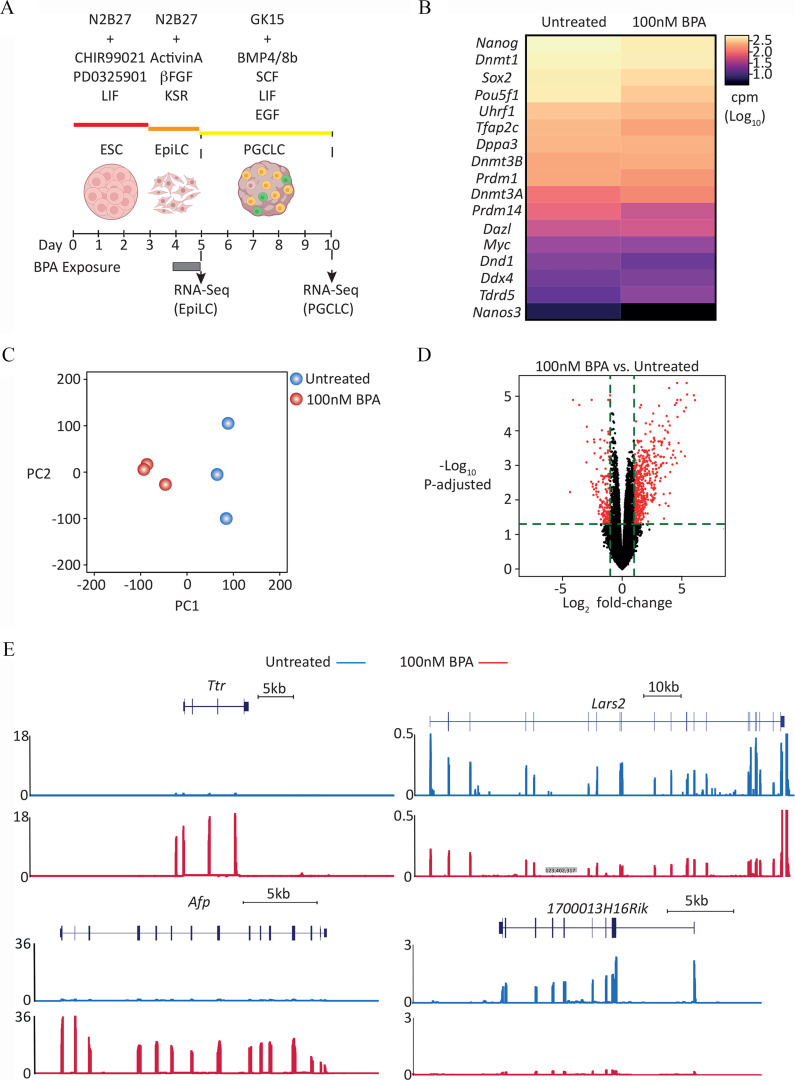
Figure 3.
Transcriptome analysis of PGCLCs derived from EpiLCs exposed to BPA. (A) Graphic showing dosing and differentiation strategy followed by cell collection for RNA-seq library generation. Uncolored cells represent Blimp1-; Stella- /DN/nongerm cells; yellow cells represent transitioning germ cells; green cells represent PGCLCs. [Illustration in part created with ©BioRender – (biorender.com), per the Biorender terms and conditions.] (B) level-ranked heatmap of germ cell markers indicated in RNA-seq data generated from d5 aggregate FACS-purified DP/PGCLCs, derived from EpiLCs exposed to control (untreated) or BPA. Candidate germ cell marker list taken from Hayashi et al. (Hayashi et al. 2011). (C) PCA of transcriptome of d5 DP/BVSC cells, derived from untreated or BPA-exposed EpiLCs. (D) Volcano plot of RNA-seq data displaying gene expression pattern of d5 DP/PGCLCs, derived from untreated or 100 nM BPA-exposed EpiLCs. Significant differentially expressed genes (, Benjamini-Hochberg–adjusted ) are highlighted in red. (E) Genome browser view of expression data for representative genes, up- (Ttr and Afp) and down-regulated (Lars2 and 1700013H16Rik) in d5 DP/BVSC cells, derived from BPA-exposed EpiLCs. Y-axis values are normalized cpm/. For each locus, top panel represents “Untreated” and bottom panel represents “ BPA exposed.” For full list of differentially up- and down-regulated genes, see Excel Tables S14 and S15, respectively. Note: , basic fibroblast growth factor; BMP4, bone morphogenetic protein 4; BMP8b, bone morphogenetic protein 8b; BPA, bisphenol A; BVSC, B lymphocyte-induced maturation protein 1 Blimp1-mVenus and stella-ECFP reporter transgenes; cpm, counts per million; DN, double negative; DP, double positive; EGF, epidermal growth factor; embryonic stem cell; EpiLC, epiblast-like cell; FACS, fluorescence-activated cell sorting; KSR, knockout serum replacement; LIF, leukemia inhibitory factor; PGC, primordial germ cell; PGCLC, PGC-like cell; SCF, stem cell factor.