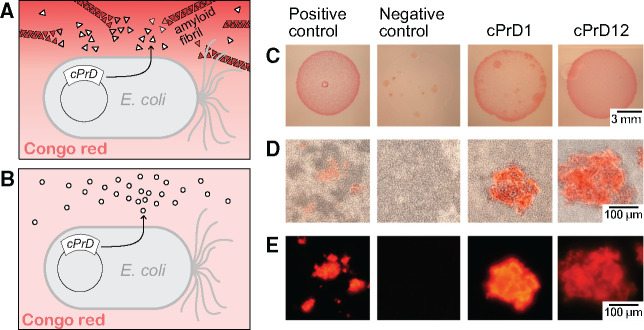
Fig. 4.
Comparison of colonies and corresponding protein aggregates produced by bacteria transformed with plasmids encoding cPrDs. (A and B) Two variants of possible results described in a cartoon. (A) Candidate cPrD forms amyloid aggregates that bind Congo red. (B) cPrD does not form amyloid aggregates therefore does not bind Congo red. (C) Colonies of Escherichia coli grew on the induction medium containing 0.1% Congo red. The red color of the colony indicates that the exported protein binds to Congo red. Binding of Congo red is typical for amyloid fibrils. (D) Light microscopy of preparations made of the same bacterial colonies show aggregates binding Congo red and bacterial cells concentrating around them. (E) Fluorescent microscopy of protein aggregates from panel (B) is shown. The amyloidogenic fragment of Sup35 (residues 2–253) was used as a positive control. Fragment of Sup35 not able to form amyloids (residues 125–253) was used as a negative control. Protein names and organisms from which candidate prion domains (cPrDs) were selected are listed in table 1.