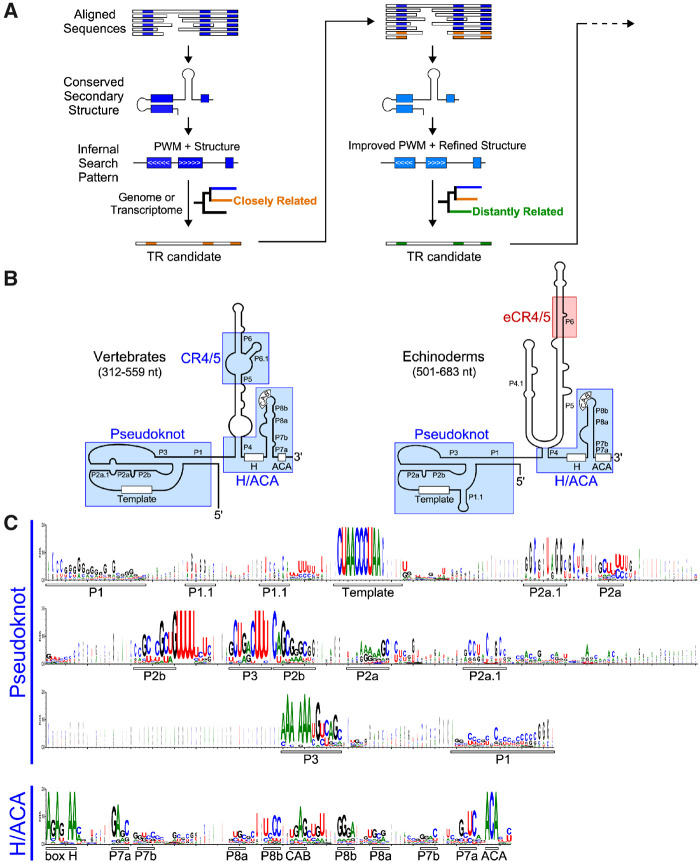
Fig. 1.
A phylogeny-assisted approach for TR identification. (A) The workflow of phylogeny-assisted reiterative homology search for novel TRs. An initial PWM and secondary structure pattern derived from known TRs was used to search for novel TR candidates in the genomes or transcriptomes of closely related species. The newly identified TR sequences were used to improve the PWM and structural pattern for searching TR candidates in distantly related species. (B) Comparison of vertebrate and echinoderm TR secondary structures. Two structural domains, T-PK and box H/ACA domains, are conserved in both vertebrate and echinoderm TRs, whereas the vertebrate CR4/5 domain that contains P6.1 stem-loop is replaced with a functionally equivalent eCR4/5 domain in echinoderms. The size ranges of vertebrate and echinoderm TRs are shown. (C) PWMs of TR sequences of the pseudoknot and H/ACA domains. PWMs are derived from the multiple-sequence alignment of 42 vertebrate TRs (Chen et al. 2000) and 13 echinoderm TRs (Li et al. 2013; Podlevsky et al. 2016b). Conserved sequence motifs and base-paired regions within each domain are indicated underneath the matrices