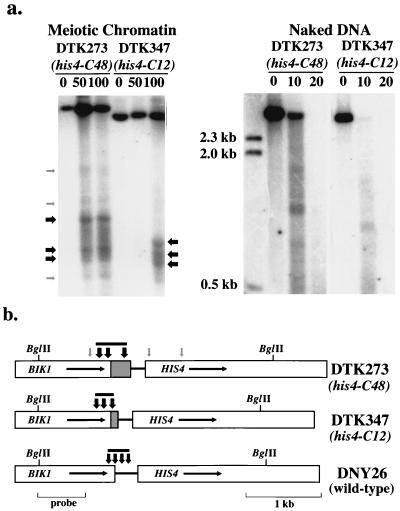
FIG. 2.
Mapping of the DNase I-hypersensitive sites in HIS4 chromatin isolated from strains DTK273 (homozygous for his4-C48) and DTK347 (homozygous for his4-C12). (a) Chromatin was prepared from cells incubated for 6 h at 25°C in sporulation medium (9). Chromatin was digested with 0, 50, and 100 U of DNase I, and DNA was then extracted. Naked DNA was incubated with 0, 10, or 20 U of DNase I. Samples were then treated with BglII and examined by Southern analysis; a BglII-PvuII fragment of BIK1 was used as a hybridization probe. Numbers on the left indicate the sizes of DNA markers. Arrows mark the positions of DNase I-hypersensitive sites; black and gray arrows indicate strong and weak sites, respectively. (b) The positions of the DNase I-hypersensitive sites relative to restriction maps for the strains are shown by vertical arrows. As above, black and gray arrows indicate strong and weak hypersensitive sites, respectively. The gray bars above the arrows indicate the extent of the major open chromatin region in each strain. The promoter region in DNY26 (wild type) is replaced by the (CCGNN)48 sequence (hatched region) in DTK273 and the (CCGNN)12 sequence in DTK347. Data for DNY26 is derived from Fan and Petes (9).