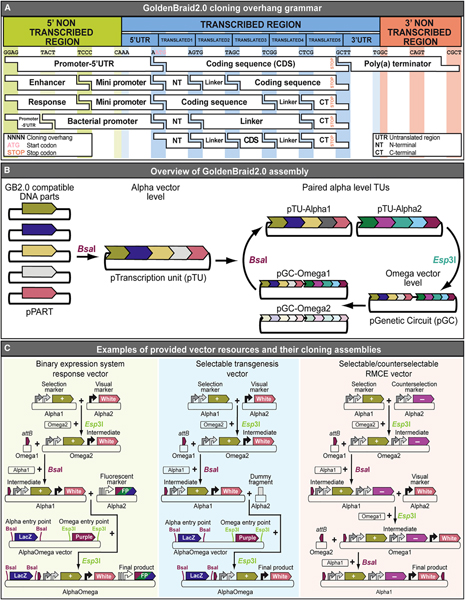
Figure 6. A selection and counterselection vector library resource for iterative synthetic assembly.
(A) Iterative synthetic assembly with Goldenbraid 2.0 (GB2.0) cloning uses a predefined “grammar” of four nucleotide overhangs that determine DNA element identity and assembly order during cloning.
(B) GB2.0 uses a one-pot assembly of multiple DNA parts into one of two alpha-level destination vectors resulting in a transcription unit (TU). Paired TUs can be further assembled into an omega-level destination vector producing a multigenic genetic circuit (GC). These can then be further assembled back into an alpha-level vector to produce ever more complex constructs.
(C) A number of pre-built selectable and/or counterselectable vectors are provided for a variety of applications as well as a library of GB2.0-compatible DNA elements totaling 121 plasmids. Each of these vectors was itself cloned from smaller elements using GB2.0 synthetic assembly. The vectors require minimal modification using either GB2.0 or traditional cut-and-paste cloning for a particular application. More experienced users can generate fully customized vectors appropriate to their needs using GB2.0 cloning. Three examples of vector resources and their cloning assemblies are shown. All plasmids are publicly available via Addgene (https://www.addgene.org/), and a full list can be found in Tables S1 and S2.
See also Figure S10.