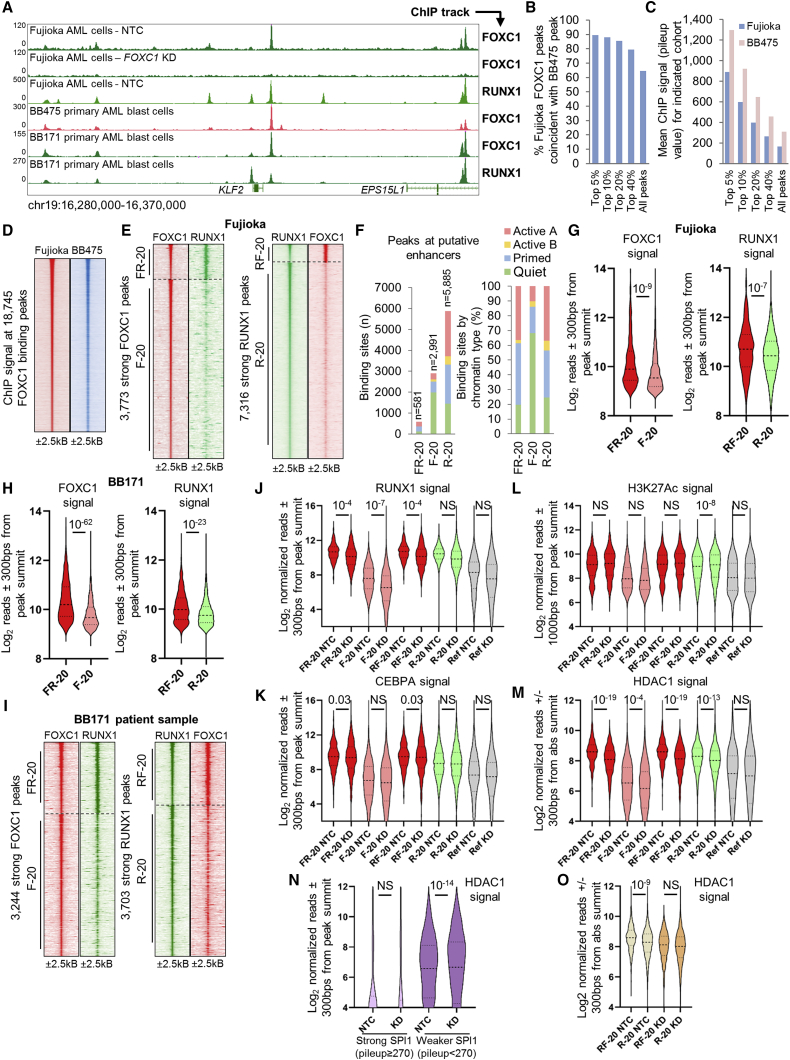
Figure 4.
FOXC1 chromatin binding in AML cells and colocalization with RUNX1
(A) Exemplar ChIP-seq tracks.
(B) Bar chart shows percentage of FOXC1 binding peaks in Fujioka AML cells in the indicated peak cohorts, which are coincident with a FOXC1 binding peak in BB475 primary AML cells.
(C) Bar chart shows mean FOXC1 ChIP signal in the indicated peak cohorts in Fujioka cells and at the same genomic locations in BB475 primary AML cells.
(D) Heatmaps show FOXC1 ChIP signal at FOXC1 binding sites in Fujioka and at the same genomic locations in BB475 primary AML cells, ranked by peak strength.
(E and I) Heatmaps show ChIP signal for RUNX1 at strong FOXC1 binding sites (left panel) and FOXC1 at strong RUNX1 binding sites (right panel) in Fujioka cells (E) or BB171 primary patient AML blast cells (I).
(F) Bar charts show chromatin categories for the indicated classes of strong FOXC1 and RUNX1 binding peaks in Fujioka cells by number (left panel) and proportion (right panel).
(G, H, and J–O) Violin plots show distribution, median (thick dotted line), and interquartile range (light dotted lines) for ChIP signal for the indicated proteins at sites with strong FOXC1 and RUNX1 binding (FR-20, FOXC1 centered; RF-20 RUNX1 centered), strong FOXC1 binding (F-20), or strong RUNX1 binding (R-20) in Fujioka AML cells (G, J–M, and O), BB171 primary patient AML blast cells (H), or SPI1 binding sites (N) for, where indicated, control cells (NTC) or following FOXC1 KD.
NS, not significant; Ref, reference cohort used for normalization between experiments. p values, unpaired t test. See also Figure S4.