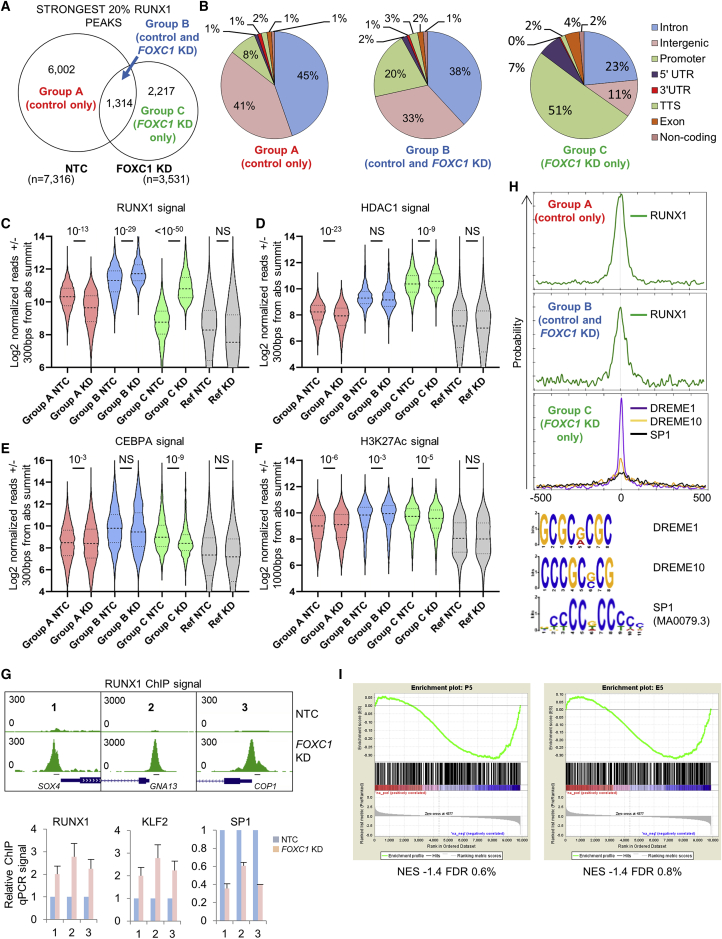
Figure 6.
FOXC1 knockdown triggers redistribution of RUNX1 binding
(A) Venn diagram shows intersection of the strongest 20% of RUNX1 binding peaks in control (NTC) or FOXC1 KD Fujioka AML cells and classification of groups.
(B) Pie charts show genome annotations for RUNX1 binding peaks in groups A–C.
(C–F) Violin plots show distribution, median (thick dotted line), and interquartile range (light dotted lines) for ChIP signal for the indicated proteins and groups in control (NTC) or FOXC1 KD Fujioka AML cells. p values, unpaired t test.
(G) Exemplar ChIP-seq tracks (upper panel) with confirmatory ChIP-PCR (lower panels; n = 3).
(H) MEME-ChIP and DREME motif enrichment plots for the indicated groups.
(I) GSEA plots.
E5, enhancers that exhibit ≥5-fold increase in RUNX1 ChIP signal; P5, genes whose promoters exhibit ≥5-fold increase in RUNX1 ChIP signal. See also Figure S6.