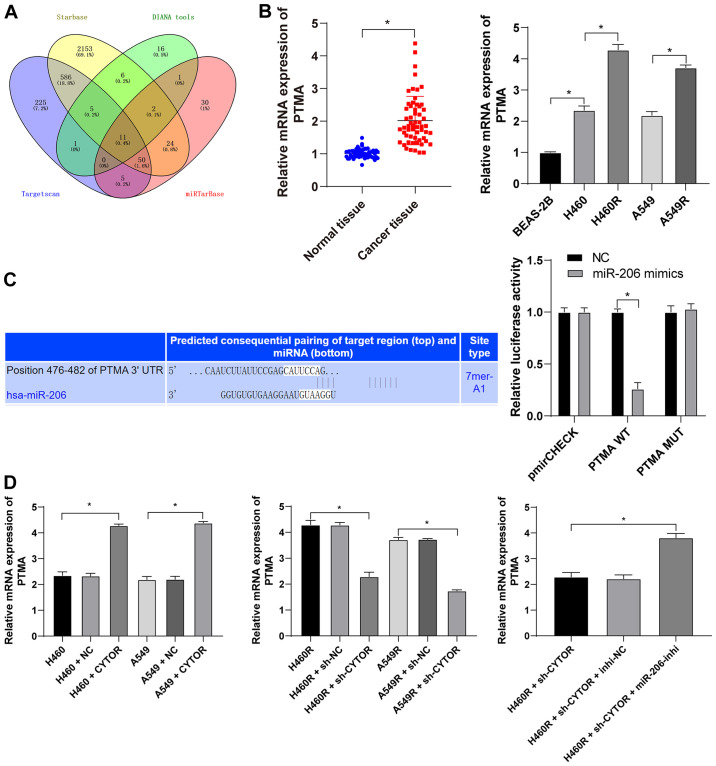
Figure 9.
miR-206 targets PTMA expression. (A) Target genes downstream of miR-206 were analyzed using various databases and their overlapping results were presented in a Venn diagram, with yellow representing starBase (http://starbase.sysu.edu.cn/), green representing DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted), blue representing TargetScan (http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg) and red representing miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php). (B) Level of PTMA mRNA in 58 pairs of non-small cell lung cancer tissues and cell lines as detected by RT-qPCR. (C) The binding site between miR-206 and PTMA was predicted via starBase (http://starbase.sysu.edu.cn/) and their binding interaction was verified by a dual-luciferase reporter assay. (D) The level of PTMA mRNA in differently treated cells as verified by RT-qPCR. Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; PTMA, prothymosin α; MUT, mutant; WT, wild-type; hsa, Homo sapiens; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; inhi, inhibitor; miR, microRNA; H460R, H460 cells with radioresistance.