Figure 4:
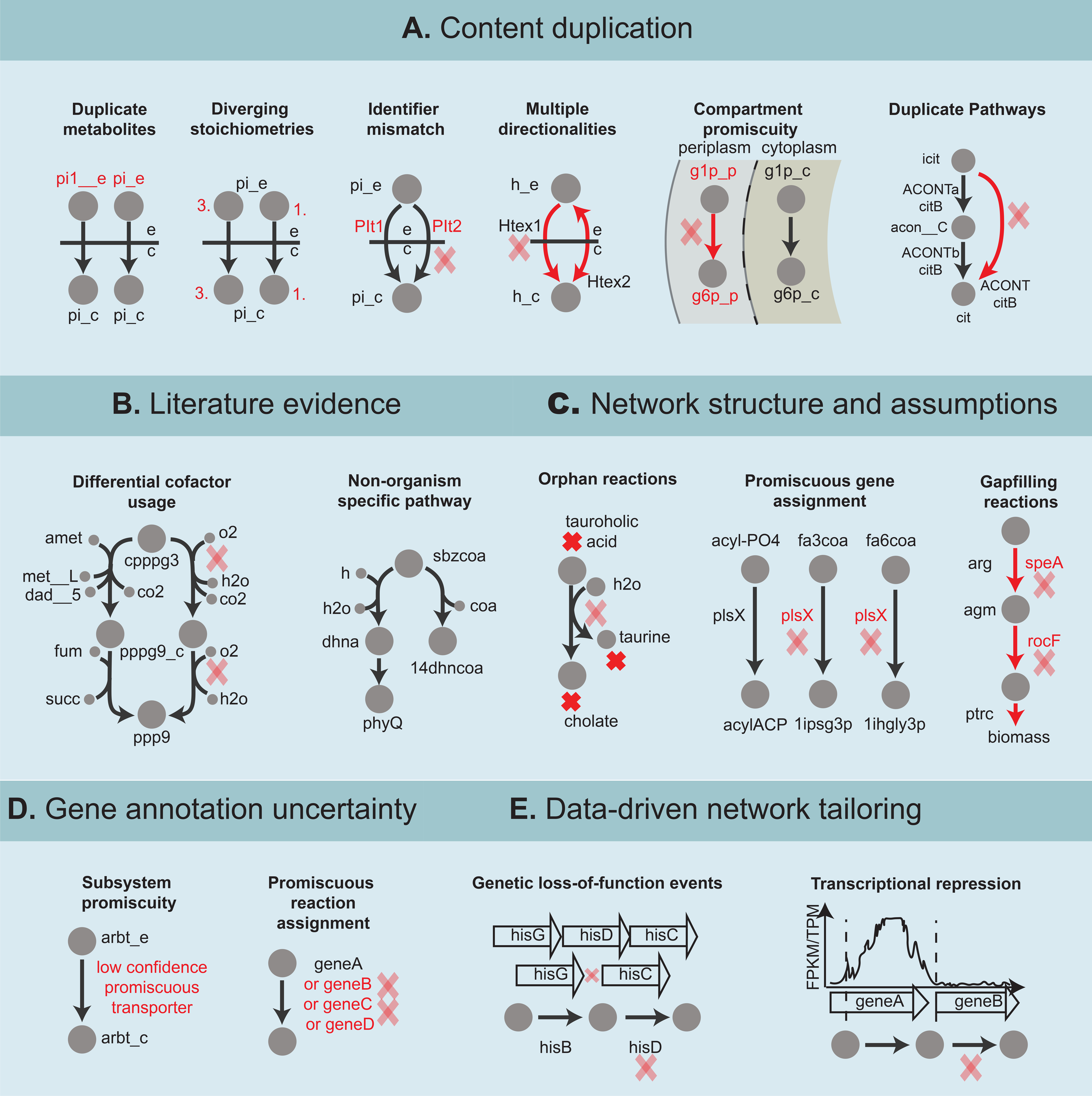
Types of content removal can be subdivided into: A) content duplication. From left to right duplicate reaction caused by; inconsistent metabolite identifier (pi_e and pi__e identify extracellular phosphate), diverging stoichiometries (three phosphate molecules transported to the cytoplasm versus one phosphate molecule transported), reaction identifier mismatch (Plt1 and Plt2 are both the same transport reaction), multiple directionalities (bi-directional transport versus import of hydrogen across the outer membrane), compartment promiscuity (erroneous assignment of a phosphoglucomutase to the periplasmic compartment converting D-glucose1phosphate to D-glucose-6-phosphate), and duplicate pathways (conversion of isocitrate to citrate via a two-step pathway and via a lumped reaction). N.B., these cases serve as examples of possible content duplication but should be checked against the literature. For example, an organism could encode two phosphate transporters both of which catalyze a transport process but with different stoichiometry. B) Organism nonspecific pathways; left: example of a heme pathway initially added to the S. aureus GEM with incorrect differential cofactor usage due to lack of knowledge and subsequently corrected as a result of recent discoveries (Lobo et al., 2015; Seif, Monk, Mih, et al., 2019), right: example of a plant pathway for vitamin K1 (phylloquinone) biosynthesis which was added to the GEM of a gram-positive bacterium likely due to insufficient curation. C) Low confidence modeling assumptions. From left to right; orphan taurocholate amidohydrolase reaction annotated as being catalyzed by a C59 family penicillin amidase, promiscuous assignment of plsX as a phosphate acyltransferase, a isohexadecanoylglycerol-3-phosphate O-acyltransferase and an isopentadecanoyl-glycerol-3-phosphate O-acyltransferase (among others), gap filled putrescine biosynthesis pathway with erroneous gene assignment. D) Low confidence gene annotation; transport reaction assigned to a low confidence transporter, generic reaction with long “OR” based gene reaction rule, E) Datadriven evidence. Reaction catalyzed by a gene carrying a loss-of-function mutation, and reaction catalyzed by a gene that is not expressed in a cell. Abbreviations: X_c = cytoplasmic, X_e = extracellular, X_p = periplasmic, h = hydrogen, pi = phosphate, icit = isocitrate, acon__C = cis-aconitate, cit = citrate, cpppg3 = coproporphyrinogen III, amet = S-adenosyl-L-methionine, met__L = L-methionine, fum = fumarate, succ = succinate, pppg9 = protoporphyrinogen IX, o2 = oxygen, h2o = water, co2 = carbon dioxide, ppp9 = protoporphyrin, sbzcoa = O-succinylbenzoylcoA, dhna = 1,4-dihydroxy-2-naphthoate, phyQ = phylloquinone, 14dhncoa = 1,4-dihydroxy-2napthoyl-coA, fa3coa = fatty acid (Iso-C15:0)-coA, fa3coa = fatty acid (Iso-C16:0)-coA, 1ipsg3p = 1-isopentadecanoyl-sn-glycerol 3-phosphate, 1ihgly3p = 1-isohexadecanoyl-sn-glycerol_3phosphate, arg = L-arginine, agm = agmatine, ptrc = putrescine, arbt = arbutin
